 A brand new learn about via 279 scientists, led via the Royal Botanic Gardens, Kew, has up to date our working out of the flowering plant tree of existence via examining genetic knowledge from over 9,500 species. This analysis, involving a global collaboration and critical technological developments, supplies a very powerful insights for plant classification, conservation, and medicinal discovery. The findings are freely available, promising to give a boost to long term botanical research and programs.Scientists have built a groundbreaking tree of existence the use of 1.8 billion letters of genetic code.A up to date learn about revealed within the magazine Nature via a global group of 279 scientists, together with 3 biologists from the College of Michigan, supplies the newest insights into the flowering plant tree of existence.The use of 1.8 billion letters of genetic code from greater than 9,500 species overlaying virtually 8,000 recognized flowering plant genera (ca. 60%), this success sheds new mild at the evolutionary historical past of flowering crops and their upward push to ecological dominance on Earth.Led via scientists on the Royal Botanic Gardens, Kew, the analysis group believes the information will help long term makes an attempt to spot new species, refine plant classification, discover new medicinal compounds, and preserve crops within the face of local weather exchange and biodiversity loss.The key milestone for plant science, involving 138 organizations the world over, used to be constructed on 15 instances extra knowledge than any related research of the flowering plant tree of existence. Some of the species sequenced for this learn about, greater than 800 have by no means had their DNA sequenced prior to.Technological Demanding situations and SolutionsThe sheer quantity of knowledge unlocked via this analysis, which might take a unmarried pc 18 years to procedure, is a large stride towards development a tree of existence for all 330,000 recognized species of flowering crops—a large enterprise byKew’s Tree of Lifestyles Initiative.“Inspecting this exceptional quantity of knowledge to decode the guidelines hidden in tens of millions of DNA sequences used to be an enormous problem. However it additionally presented the original alternative to reevaluate and lengthen our wisdom of the plant tree of existence, opening a brand new window to discover the complexity of plant evolution,” mentioned Alexandre Zuntini, a analysis fellow at Royal Botanic Gardens, Kew.Tom Carruthers, postdoctoral researcher within the lab of U-M evolutionary biologist Stephen Smith, is co-lead writer of the learn about with Zuntini, who he up to now labored with at Kew. U-M plant systematist Richard Rabeler is a co-author.
A brand new learn about via 279 scientists, led via the Royal Botanic Gardens, Kew, has up to date our working out of the flowering plant tree of existence via examining genetic knowledge from over 9,500 species. This analysis, involving a global collaboration and critical technological developments, supplies a very powerful insights for plant classification, conservation, and medicinal discovery. The findings are freely available, promising to give a boost to long term botanical research and programs.Scientists have built a groundbreaking tree of existence the use of 1.8 billion letters of genetic code.A up to date learn about revealed within the magazine Nature via a global group of 279 scientists, together with 3 biologists from the College of Michigan, supplies the newest insights into the flowering plant tree of existence.The use of 1.8 billion letters of genetic code from greater than 9,500 species overlaying virtually 8,000 recognized flowering plant genera (ca. 60%), this success sheds new mild at the evolutionary historical past of flowering crops and their upward push to ecological dominance on Earth.Led via scientists on the Royal Botanic Gardens, Kew, the analysis group believes the information will help long term makes an attempt to spot new species, refine plant classification, discover new medicinal compounds, and preserve crops within the face of local weather exchange and biodiversity loss.The key milestone for plant science, involving 138 organizations the world over, used to be constructed on 15 instances extra knowledge than any related research of the flowering plant tree of existence. Some of the species sequenced for this learn about, greater than 800 have by no means had their DNA sequenced prior to.Technological Demanding situations and SolutionsThe sheer quantity of knowledge unlocked via this analysis, which might take a unmarried pc 18 years to procedure, is a large stride towards development a tree of existence for all 330,000 recognized species of flowering crops—a large enterprise byKew’s Tree of Lifestyles Initiative.“Inspecting this exceptional quantity of knowledge to decode the guidelines hidden in tens of millions of DNA sequences used to be an enormous problem. However it additionally presented the original alternative to reevaluate and lengthen our wisdom of the plant tree of existence, opening a brand new window to discover the complexity of plant evolution,” mentioned Alexandre Zuntini, a analysis fellow at Royal Botanic Gardens, Kew.Tom Carruthers, postdoctoral researcher within the lab of U-M evolutionary biologist Stephen Smith, is co-lead writer of the learn about with Zuntini, who he up to now labored with at Kew. U-M plant systematist Richard Rabeler is a co-author.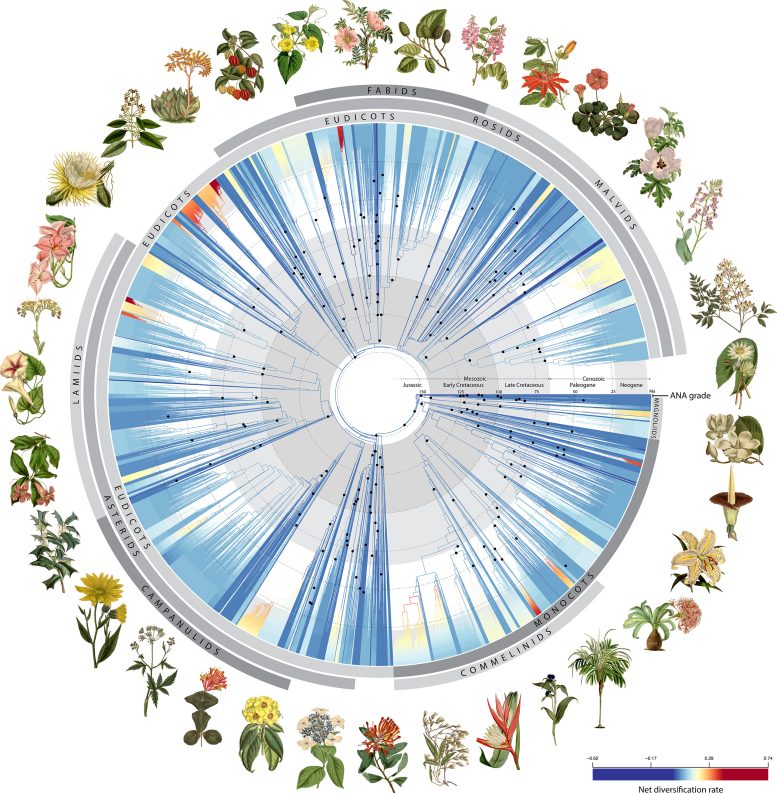 Angiosperm Tree of Lifestyles. Credit score: RBG Kew“Flowering crops feed, dress, and greet us on every occasion we stroll into the woods. The development of a flowering plant tree of existence has been a vital problem and function for the sector of evolutionary biology for greater than a century,” mentioned Smith, co-author of the learn about and professor within the U-M Division of Ecology and Evolutionary Biology. “This undertaking strikes us nearer to that function via offering a large dataset for lots of the genera of flowering crops and providing one option to whole this function.”Smith had two roles at the undertaking. First, participants of his lab—together with former U-M graduate scholar Drew Larson—traveled to Kew to assist collection participants of a big and numerous plant workforce referred to as Ericales, which contains blueberries, tea, ebony, azaleas, rhododendrons and Brazil nuts.2nd, Smith supervised the analyses and development of the undertaking dataset at the side of William Baker and Felix Woodland of the Royal Botanic Gardens, Kew, and Wolf Eisenhardt of Aarhus College.“Probably the most largest demanding situations confronted via the group used to be the surprising complexity underlying lots of the gene areas, the place other genes inform other evolutionary histories. Procedures needed to be evolved to inspect those patterns on a scale that hadn’t been accomplished prior to,” mentioned Smith, who may be director of the Program in Biology and an affiliate curator in biodiversity informatics on the U-M Herbarium.New Insights into EvolutionAs co-leader of the learn about, Carruthers’ major obligations integrated scaling the evolutionary tree to time the use of 200 fossils, examining the other evolutionary histories of the genes underlying the entire evolutionary tree, and estimating charges of diversification in numerous flowering plant lineages at other instances.“Setting up any such massive tree of existence for flowering crops, in response to such a lot of genes, sheds mild at the evolutionary historical past of this particular workforce, serving to us to know how they got here to be such an integral and dominant a part of the arena,” Carruthers mentioned. “The evolutionary relationships which are introduced—and the information underlying them—will supply a very powerful basis for numerous long term research.”The flowering plant tree of existence, similar to our personal circle of relatives tree, permits us to know how other species are comparable to one another. The tree of existence is exposed via evaluating DNA sequences between other species to spot adjustments (mutations) that gather through the years like a molecular fossil file.Our working out of the tree of existence is bettering abruptly in tandem with advances in DNA sequencing era. For this learn about, new genomic ways had been evolved to magnetically seize masses of genes and masses of hundreds of letters of genetic code from each and every pattern, orders of magnitude greater than previous strategies.
Angiosperm Tree of Lifestyles. Credit score: RBG Kew“Flowering crops feed, dress, and greet us on every occasion we stroll into the woods. The development of a flowering plant tree of existence has been a vital problem and function for the sector of evolutionary biology for greater than a century,” mentioned Smith, co-author of the learn about and professor within the U-M Division of Ecology and Evolutionary Biology. “This undertaking strikes us nearer to that function via offering a large dataset for lots of the genera of flowering crops and providing one option to whole this function.”Smith had two roles at the undertaking. First, participants of his lab—together with former U-M graduate scholar Drew Larson—traveled to Kew to assist collection participants of a big and numerous plant workforce referred to as Ericales, which contains blueberries, tea, ebony, azaleas, rhododendrons and Brazil nuts.2nd, Smith supervised the analyses and development of the undertaking dataset at the side of William Baker and Felix Woodland of the Royal Botanic Gardens, Kew, and Wolf Eisenhardt of Aarhus College.“Probably the most largest demanding situations confronted via the group used to be the surprising complexity underlying lots of the gene areas, the place other genes inform other evolutionary histories. Procedures needed to be evolved to inspect those patterns on a scale that hadn’t been accomplished prior to,” mentioned Smith, who may be director of the Program in Biology and an affiliate curator in biodiversity informatics on the U-M Herbarium.New Insights into EvolutionAs co-leader of the learn about, Carruthers’ major obligations integrated scaling the evolutionary tree to time the use of 200 fossils, examining the other evolutionary histories of the genes underlying the entire evolutionary tree, and estimating charges of diversification in numerous flowering plant lineages at other instances.“Setting up any such massive tree of existence for flowering crops, in response to such a lot of genes, sheds mild at the evolutionary historical past of this particular workforce, serving to us to know how they got here to be such an integral and dominant a part of the arena,” Carruthers mentioned. “The evolutionary relationships which are introduced—and the information underlying them—will supply a very powerful basis for numerous long term research.”The flowering plant tree of existence, similar to our personal circle of relatives tree, permits us to know how other species are comparable to one another. The tree of existence is exposed via evaluating DNA sequences between other species to spot adjustments (mutations) that gather through the years like a molecular fossil file.Our working out of the tree of existence is bettering abruptly in tandem with advances in DNA sequencing era. For this learn about, new genomic ways had been evolved to magnetically seize masses of genes and masses of hundreds of letters of genetic code from each and every pattern, orders of magnitude greater than previous strategies. Arenaria globilfora. Credit score: RBG KewA key good thing about the group’s method is that it permits a large range of plant subject material, outdated and new, to be sequenced, even if the DNA is improperly broken. The huge treasure troves of dried plant subject material on this planet’s herbarium collections, which include just about 400 million clinical specimens of crops, can now be studied genetically.“In some ways, this novel method has allowed us to collaborate with the botanists of the previous via tapping into the wealth of knowledge locked up in ancient herbarium specimens, a few of that have been gathered way back to the early nineteenth century,” mentioned Baker, senior analysis chief for Kew’s Tree of Lifestyles Initiative.“Our illustrious predecessors, similar to Charles Darwin or Joseph Hooker, may just now not have expected how essential those specimens could be in genomic analysis nowadays. DNA used to be now not even found out of their lifetimes. Our paintings displays simply how essential those implausible botanical museums are to groundbreaking research of existence on Earth. Who is aware of what different undiscovered science alternatives lie inside them?”Throughout all 9,506 species sequenced, greater than 3,400 got here from subject material sourced from 163 herbaria in 48 nations.“Sampling herbarium specimens for the learn about of plant relationships makes wide sampling from numerous spaces of the arena a lot more possible than if one needed to go back and forth to get recent subject material from the sector,” mentioned U-M’s Rabeler, a analysis scientist emeritus and previous assortment supervisor on the U-M Herbarium.For the tree of existence undertaking, Rabeler helped test the id of herbarium specimens decided on for sampling and analyzed the ensuing knowledge.Flowering crops on my own account for approximately 90% of all recognized plant existence on land and are discovered just about far and wide on the earth—from the steamiest tropics to the rocky outcrops of the Antarctic Peninsula. And but, our working out of ways those crops got here to dominate the scene quickly after their beginning has baffled scientists for generations, together with Darwin.Flowering crops originated greater than 140 million years in the past and then they abruptly overtook different vascular crops together with their closest residing family members—the gymnosperms (nonflowering crops that experience bare seeds, similar to cycads, conifers, and ginkgo).Darwin used to be mystified via the apparently surprising look of such range within the fossil file. In an 1879 letter to Hooker, his shut confidant and director of the Royal Botanic Gardens, Kew, he wrote: “The fast construction so far as we will be able to pass judgement on of all of the upper crops inside contemporary geological instances is an abominable thriller.”The use of 200 fossils, the authors scaled their tree of existence to time, revealing how flowering crops advanced throughout geological time. They discovered that early flowering crops did certainly explode in range, giving upward push to greater than 80% of the most important lineages that exist nowadays in a while after their beginning.On the other hand, this pattern then declined to a steadier fee for the following 100 million years till any other surge in diversification about 40 million years in the past, coinciding with an international decline in temperatures. Those new insights would have fascinated Darwin and can indubitably assist nowadays’s scientists grappling with the demanding situations of working out how and why species diversify.World Collaboration and Open AccessAssembling a tree of existence this in depth would had been not possible with out Kew’s scientists taking part with many companions around the globe. In general, 279 authors had been concerned within the analysis, representing many various nationalities from 138 organizations in 27 nations.“The plant neighborhood has an extended historical past of taking part and coordinating molecular sequencing to generate a extra complete and strong plant tree of existence. The hassle that ended in this paper continues in that custom however scales up somewhat considerably,” mentioned U-M’s Smith.The flowering plant tree of existence has monumental doable in biodiversity analysis. It is because, simply as one can expect the homes of a component in response to its place within the periodic desk, the positioning of a species within the tree of existence lets in us to expect its homes. The brand new knowledge will thus be priceless for reinforcing many spaces of science and past.To allow this, the tree and all the knowledge that underpin it had been made brazenly and freely available to each the general public and clinical neighborhood, together with thru theKew Tree of Lifestyles Explorer.Open get admission to will assist scientists to make the most productive use of the information, similar to combining it with synthetic intelligence to expect which plant species might come with molecules with medicinal doable.In a similar fashion, the tree of existence can be utilized to raised perceive and expect how pests and sicknesses are going to impact crops sooner or later. In the long run, the authors word, the programs of this knowledge might be pushed via the ingenuity of the scientists having access to it.Reference: “Phylogenomics and the upward push of the angiosperms” via Alexandre R. Zuntini, Tom Carruthers, Olivier Maurin, Paul C. Bailey, Kevin Leempoel, Grace E. Brewer, Niroshini Epitawalage, Elaine Françoso, Berta Gallego-Paramo, Catherine McGinnie, Raquel Negrão, Shyamali R. Roy, Lalita Simpson, Eduardo Toledo Romero, Vanessa M. A. Barber, Laura Botigué, James J. Clarkson, Robyn S. Cowan, Steven Dodsworth, Matthew G. Johnson, Jan T. Kim, Lisa Pokorny, Norman J. Wickett, Guilherme M. Antar, Lucinda DeBolt, Karime Gutierrez, Kasper P. Hendriks, Alina Hoewener, Ai-Qun Hu, Elizabeth M. Joyce, Izai A. B. S. Kikuchi, Isabel Larridon, Drew A. Larson, Elton John de Lírio, Jing-Xia Liu, Panagiota Malakasi, Natalia A. S. Przelomska, Toral Shah, Juan Viruel, Theodore R. Allnutt, Gabriel Okay. Ameka, Rose L. Andrew, Marc S. Appelhans, Montserrat Arista, María Jesús Ariza, Juan Arroyo, Watchara Arthan, Julien B. Bachelier, C. Donovan Bailey, Helen F. Barnes, Matthew D. Barrett, Russell L. Barrett, Randall J. Bayer, Michael J. Bayly, Ed Biffin, Nicky Biggs, Joanne L. Birch, Diego Bogarín, Renata Borosova, Alexander M. C. Bowles, Peter C. Boyce, Gemma L. C. Bramley, Marie Briggs, Linda Broadhurst, Gillian Okay. Brown, Jeremy J. Bruhl, Anne Bruneau, Sven Buerki, Edie Burns, Margaret Byrne, Stuart Cable, Ainsley Calladine, Martin W. Callmander, Ángela Cano, David J. Cantrill, Warren M. Cardinal-McTeague, Mónica M. Carlsen, Abigail J. A. Carruthers, Alejandra de Castro Mateo, Mark W. Chase, Lars W. Chatrou, Martin Cheek, Shilin Chen, Maarten J. M. Christenhusz, Pascal-Antoine Christin, Mark A. Clements, Skye C. Coffey, John G. Conran, Xavier Cornejo, Thomas L. P. Couvreur, Ian D. Cowie, Laszlo Csiba, Iain Darbyshire, Gerrit Davidse, Nina M. J. Davies, Aaron P. Davis, Kor-jent van Dijk, Stephen R. Downie, Marco F. Duretto, Melvin R. Duvall, Sara L. Edwards, Urs Eggli, Roy H. J. Erkens, Marcial Escudero, Manuel de los angeles Estrella, Federico Fabriani, Michael F. Fay, Paola de L. Ferreira, Sarah Z. Ficinski, Rachael M. Fowler, Sue Frisby, Lin Fu, Tim Fulcher, Mercè Galbany-Casals, Elliot M. Gardner, Dmitry A. German, Augusto Giaretta, Marc Gibernau, Lynn J. Gillespie, Cynthia C. González, David J. Goyder, Sean W. Graham, Aurélie Grall, Laura Inexperienced, Bee F. Gunn, Diego G. Gutiérrez, Jan Hackel, Thomas Haevermans, Anna Haigh, Jocelyn C. Corridor, Tony Corridor, Melissa J. Harrison, Sebastian A. Hatt, Oriane Hidalgo, Trevor R. Hodkinson, Gareth D. Holmes, Helen C. F. Hopkins, Christopher J. Jackson, Shelley A. James, Richard W. Jobson, Gudrun Kadereit, Imalka M. Kahandawala, Kent Kainulainen, Masahiro Kato, Elizabeth A. Kellogg, Graham J. King, Beata Klejevskaja, Bente B. Klitgaard, Ronell R. Klopper, Sandra Knapp, Marcus A. Koch, James H. Leebens-Mack, Frederic Lens, Christine J. Leon, Étienne Léveillé-Bourret, Gwilym P. Lewis, De-Zhu Li, Lan Li, Sigrid Liede-Schumann, Tatyana Livshultz, David Lorence, Meng Lu, Patricia Lu-Irving, Jaquelini Luber, Eve J. Lucas, Manuel Luján, Mabel Lum, Terry D. Macfarlane, Carlos Magdalena, Vidal F. Mansano, Lizo E. Masters, Simon J. Mayo, Kristina McColl, Angela J. McDonnell, Andrew E. McDougall, Todd G. B. McLay, Hannah McPherson, Rosa I. Meneses, Vincent S. F. T. Merckx, Fabián A. Michelangeli, John D. Mitchell, Alexandre Okay. Monro, Michael J. Moore, Taryn L. Mueller, Klaus Mummenhoff, Jérôme Munzinger, Priscilla Muriel, Daniel J. Murphy, Katharina Nargar, Lars Nauheimer, Francis J. Nge, Reto Nyffeler, Andrés Orejuela, Edgardo M. Ortiz, Luis Palazzesi, Ariane Luna Peixoto, Susan Okay. Pell, Jaume Pellicer, Darin S. Penneys, Oscar A. Perez-Escobar, Claes Persson, Marc Pignal, Yohan Pillon, José R. Pirani, Gregory M. Plunkett, Robyn F. Powell, Ghillean T. Prance, Carmen Puglisi, Ming Qin, Richard Okay. Rabeler, Paul E. J. Rees, Matthew Renner, Eric H. Roalson, Michele Rodda, Zachary S. Rogers, Saba Rokni, Rolf Rutishauser, Miguel F. de Salas, Hanno Schaefer, Rowan J. Schley, Alexander Schmidt-Lebuhn, Alison Shapcott, Ihsan Al-Shehbaz, Kelly A. Shepherd, Mark P. Simmons, André O. Simões, Ana Rita G. Simões, Michelle Siros, Eric C. Smidt, James F. Smith, Neil Snow, Douglas E. Soltis, Pamela S. Soltis, Robert J. Soreng, Cynthia A. Sothers, Julian R. Starr, Peter F. Stevens, Shannon C. Okay. Straub, Lena Struwe, Jennifer M. Taylor, Ian R. H. Telford, Andrew H. Thornhill, Ifeanna Enamel, Anna Trias-Blasi, Frank Udovicic, Timothy M. A. Utteridge, Jose C. Del Valle, G. Anthony Verboom, Helen P. Vonow, Maria S. Vorontsova, Jurriaan M. de Vos, Noor Al-Wattar, Michelle Waycott, Cassiano A. D. Welker, Adam J. White, Jan J. Wieringa, Luis T. Williamson, Trevor C. Wilson, Sin Yeng Wong, Lisa A. Woods, Roseina Woods, Stuart Worboys, Martin Xanthos, Ya Yang, Yu-Xiao Zhang, Meng-Yuan Zhou, Sue Zmarzty, Fernando O. Zuloaga, Alexandre Antonelli, Sidonie Bellot, Darren M. Crayn, Olwen M. Grace, Paul J. Kersey, Ilia J. Leitch, Hervé Sauquet, Stephen A. Smith, Wolf L. Eiserhardt, Félix Woodland and William J. Baker, 24 April 2024, Nature.
Arenaria globilfora. Credit score: RBG KewA key good thing about the group’s method is that it permits a large range of plant subject material, outdated and new, to be sequenced, even if the DNA is improperly broken. The huge treasure troves of dried plant subject material on this planet’s herbarium collections, which include just about 400 million clinical specimens of crops, can now be studied genetically.“In some ways, this novel method has allowed us to collaborate with the botanists of the previous via tapping into the wealth of knowledge locked up in ancient herbarium specimens, a few of that have been gathered way back to the early nineteenth century,” mentioned Baker, senior analysis chief for Kew’s Tree of Lifestyles Initiative.“Our illustrious predecessors, similar to Charles Darwin or Joseph Hooker, may just now not have expected how essential those specimens could be in genomic analysis nowadays. DNA used to be now not even found out of their lifetimes. Our paintings displays simply how essential those implausible botanical museums are to groundbreaking research of existence on Earth. Who is aware of what different undiscovered science alternatives lie inside them?”Throughout all 9,506 species sequenced, greater than 3,400 got here from subject material sourced from 163 herbaria in 48 nations.“Sampling herbarium specimens for the learn about of plant relationships makes wide sampling from numerous spaces of the arena a lot more possible than if one needed to go back and forth to get recent subject material from the sector,” mentioned U-M’s Rabeler, a analysis scientist emeritus and previous assortment supervisor on the U-M Herbarium.For the tree of existence undertaking, Rabeler helped test the id of herbarium specimens decided on for sampling and analyzed the ensuing knowledge.Flowering crops on my own account for approximately 90% of all recognized plant existence on land and are discovered just about far and wide on the earth—from the steamiest tropics to the rocky outcrops of the Antarctic Peninsula. And but, our working out of ways those crops got here to dominate the scene quickly after their beginning has baffled scientists for generations, together with Darwin.Flowering crops originated greater than 140 million years in the past and then they abruptly overtook different vascular crops together with their closest residing family members—the gymnosperms (nonflowering crops that experience bare seeds, similar to cycads, conifers, and ginkgo).Darwin used to be mystified via the apparently surprising look of such range within the fossil file. In an 1879 letter to Hooker, his shut confidant and director of the Royal Botanic Gardens, Kew, he wrote: “The fast construction so far as we will be able to pass judgement on of all of the upper crops inside contemporary geological instances is an abominable thriller.”The use of 200 fossils, the authors scaled their tree of existence to time, revealing how flowering crops advanced throughout geological time. They discovered that early flowering crops did certainly explode in range, giving upward push to greater than 80% of the most important lineages that exist nowadays in a while after their beginning.On the other hand, this pattern then declined to a steadier fee for the following 100 million years till any other surge in diversification about 40 million years in the past, coinciding with an international decline in temperatures. Those new insights would have fascinated Darwin and can indubitably assist nowadays’s scientists grappling with the demanding situations of working out how and why species diversify.World Collaboration and Open AccessAssembling a tree of existence this in depth would had been not possible with out Kew’s scientists taking part with many companions around the globe. In general, 279 authors had been concerned within the analysis, representing many various nationalities from 138 organizations in 27 nations.“The plant neighborhood has an extended historical past of taking part and coordinating molecular sequencing to generate a extra complete and strong plant tree of existence. The hassle that ended in this paper continues in that custom however scales up somewhat considerably,” mentioned U-M’s Smith.The flowering plant tree of existence has monumental doable in biodiversity analysis. It is because, simply as one can expect the homes of a component in response to its place within the periodic desk, the positioning of a species within the tree of existence lets in us to expect its homes. The brand new knowledge will thus be priceless for reinforcing many spaces of science and past.To allow this, the tree and all the knowledge that underpin it had been made brazenly and freely available to each the general public and clinical neighborhood, together with thru theKew Tree of Lifestyles Explorer.Open get admission to will assist scientists to make the most productive use of the information, similar to combining it with synthetic intelligence to expect which plant species might come with molecules with medicinal doable.In a similar fashion, the tree of existence can be utilized to raised perceive and expect how pests and sicknesses are going to impact crops sooner or later. In the long run, the authors word, the programs of this knowledge might be pushed via the ingenuity of the scientists having access to it.Reference: “Phylogenomics and the upward push of the angiosperms” via Alexandre R. Zuntini, Tom Carruthers, Olivier Maurin, Paul C. Bailey, Kevin Leempoel, Grace E. Brewer, Niroshini Epitawalage, Elaine Françoso, Berta Gallego-Paramo, Catherine McGinnie, Raquel Negrão, Shyamali R. Roy, Lalita Simpson, Eduardo Toledo Romero, Vanessa M. A. Barber, Laura Botigué, James J. Clarkson, Robyn S. Cowan, Steven Dodsworth, Matthew G. Johnson, Jan T. Kim, Lisa Pokorny, Norman J. Wickett, Guilherme M. Antar, Lucinda DeBolt, Karime Gutierrez, Kasper P. Hendriks, Alina Hoewener, Ai-Qun Hu, Elizabeth M. Joyce, Izai A. B. S. Kikuchi, Isabel Larridon, Drew A. Larson, Elton John de Lírio, Jing-Xia Liu, Panagiota Malakasi, Natalia A. S. Przelomska, Toral Shah, Juan Viruel, Theodore R. Allnutt, Gabriel Okay. Ameka, Rose L. Andrew, Marc S. Appelhans, Montserrat Arista, María Jesús Ariza, Juan Arroyo, Watchara Arthan, Julien B. Bachelier, C. Donovan Bailey, Helen F. Barnes, Matthew D. Barrett, Russell L. Barrett, Randall J. Bayer, Michael J. Bayly, Ed Biffin, Nicky Biggs, Joanne L. Birch, Diego Bogarín, Renata Borosova, Alexander M. C. Bowles, Peter C. Boyce, Gemma L. C. Bramley, Marie Briggs, Linda Broadhurst, Gillian Okay. Brown, Jeremy J. Bruhl, Anne Bruneau, Sven Buerki, Edie Burns, Margaret Byrne, Stuart Cable, Ainsley Calladine, Martin W. Callmander, Ángela Cano, David J. Cantrill, Warren M. Cardinal-McTeague, Mónica M. Carlsen, Abigail J. A. Carruthers, Alejandra de Castro Mateo, Mark W. Chase, Lars W. Chatrou, Martin Cheek, Shilin Chen, Maarten J. M. Christenhusz, Pascal-Antoine Christin, Mark A. Clements, Skye C. Coffey, John G. Conran, Xavier Cornejo, Thomas L. P. Couvreur, Ian D. Cowie, Laszlo Csiba, Iain Darbyshire, Gerrit Davidse, Nina M. J. Davies, Aaron P. Davis, Kor-jent van Dijk, Stephen R. Downie, Marco F. Duretto, Melvin R. Duvall, Sara L. Edwards, Urs Eggli, Roy H. J. Erkens, Marcial Escudero, Manuel de los angeles Estrella, Federico Fabriani, Michael F. Fay, Paola de L. Ferreira, Sarah Z. Ficinski, Rachael M. Fowler, Sue Frisby, Lin Fu, Tim Fulcher, Mercè Galbany-Casals, Elliot M. Gardner, Dmitry A. German, Augusto Giaretta, Marc Gibernau, Lynn J. Gillespie, Cynthia C. González, David J. Goyder, Sean W. Graham, Aurélie Grall, Laura Inexperienced, Bee F. Gunn, Diego G. Gutiérrez, Jan Hackel, Thomas Haevermans, Anna Haigh, Jocelyn C. Corridor, Tony Corridor, Melissa J. Harrison, Sebastian A. Hatt, Oriane Hidalgo, Trevor R. Hodkinson, Gareth D. Holmes, Helen C. F. Hopkins, Christopher J. Jackson, Shelley A. James, Richard W. Jobson, Gudrun Kadereit, Imalka M. Kahandawala, Kent Kainulainen, Masahiro Kato, Elizabeth A. Kellogg, Graham J. King, Beata Klejevskaja, Bente B. Klitgaard, Ronell R. Klopper, Sandra Knapp, Marcus A. Koch, James H. Leebens-Mack, Frederic Lens, Christine J. Leon, Étienne Léveillé-Bourret, Gwilym P. Lewis, De-Zhu Li, Lan Li, Sigrid Liede-Schumann, Tatyana Livshultz, David Lorence, Meng Lu, Patricia Lu-Irving, Jaquelini Luber, Eve J. Lucas, Manuel Luján, Mabel Lum, Terry D. Macfarlane, Carlos Magdalena, Vidal F. Mansano, Lizo E. Masters, Simon J. Mayo, Kristina McColl, Angela J. McDonnell, Andrew E. McDougall, Todd G. B. McLay, Hannah McPherson, Rosa I. Meneses, Vincent S. F. T. Merckx, Fabián A. Michelangeli, John D. Mitchell, Alexandre Okay. Monro, Michael J. Moore, Taryn L. Mueller, Klaus Mummenhoff, Jérôme Munzinger, Priscilla Muriel, Daniel J. Murphy, Katharina Nargar, Lars Nauheimer, Francis J. Nge, Reto Nyffeler, Andrés Orejuela, Edgardo M. Ortiz, Luis Palazzesi, Ariane Luna Peixoto, Susan Okay. Pell, Jaume Pellicer, Darin S. Penneys, Oscar A. Perez-Escobar, Claes Persson, Marc Pignal, Yohan Pillon, José R. Pirani, Gregory M. Plunkett, Robyn F. Powell, Ghillean T. Prance, Carmen Puglisi, Ming Qin, Richard Okay. Rabeler, Paul E. J. Rees, Matthew Renner, Eric H. Roalson, Michele Rodda, Zachary S. Rogers, Saba Rokni, Rolf Rutishauser, Miguel F. de Salas, Hanno Schaefer, Rowan J. Schley, Alexander Schmidt-Lebuhn, Alison Shapcott, Ihsan Al-Shehbaz, Kelly A. Shepherd, Mark P. Simmons, André O. Simões, Ana Rita G. Simões, Michelle Siros, Eric C. Smidt, James F. Smith, Neil Snow, Douglas E. Soltis, Pamela S. Soltis, Robert J. Soreng, Cynthia A. Sothers, Julian R. Starr, Peter F. Stevens, Shannon C. Okay. Straub, Lena Struwe, Jennifer M. Taylor, Ian R. H. Telford, Andrew H. Thornhill, Ifeanna Enamel, Anna Trias-Blasi, Frank Udovicic, Timothy M. A. Utteridge, Jose C. Del Valle, G. Anthony Verboom, Helen P. Vonow, Maria S. Vorontsova, Jurriaan M. de Vos, Noor Al-Wattar, Michelle Waycott, Cassiano A. D. Welker, Adam J. White, Jan J. Wieringa, Luis T. Williamson, Trevor C. Wilson, Sin Yeng Wong, Lisa A. Woods, Roseina Woods, Stuart Worboys, Martin Xanthos, Ya Yang, Yu-Xiao Zhang, Meng-Yuan Zhou, Sue Zmarzty, Fernando O. Zuloaga, Alexandre Antonelli, Sidonie Bellot, Darren M. Crayn, Olwen M. Grace, Paul J. Kersey, Ilia J. Leitch, Hervé Sauquet, Stephen A. Smith, Wolf L. Eiserhardt, Félix Woodland and William J. Baker, 24 April 2024, Nature.
DOI: 10.1038/s41586-024-07324-0
Biologists Assemble Groundbreaking Tree of Lifestyles The use of 1.8 Billion Letters of Genetic Code















