To know phenotypic variations between Omicron subvariants, and the selective forces riding their evolution, we when compared replication of, and host responses to, BA.1–BA.5 with Delta, the up to now dominant VOC, in Calu-3 human airway epithelial cells (HAEs; Fig. 1). We equalized enter dose of each and every variant by means of viral envelope (E) gene copies (quantitative opposite transcription polymerase chain response, RT–qPCR) as this guarantees cells are uncovered to equivalent beginning quantities of viral RNA, which is the main viral PAMP activating defensive host innate immune responses1,19. Most significantly, this method normalizes dose independently of variant-specific variations in mobile tropism or access routes (Fig. 1a and Prolonged Information Fig. 1a,b)20,21,22, which we and others have proven have an effect on each titre decision and enter equalization by means of cell-line infectivity measurements akin to 50% tissue tradition infectious dose (TCID50) or plaque assay (Prolonged Information Fig. 1c–e). Our method is especially related for evaluating Omicron subvariants as a result of Omicron spike mutations were proven to vary tropism, expanding cathepsin-dependent endosomal access and lowering dependence on mobile floor TMPRSS2 (refs. 20,21,22,23), regardless of virion spike cleavage potency (Prolonged Information Fig. 1f). Endosomal cathepsins or mobile floor TMPRSS2 are required to cleave spike sooner than ACE2-mediated entry24,25. Certainly, in keeping with up to now printed data20,21,22, we’ve got discovered that Omicron, in particular BA.5, has enhanced access (cathepsin dependent and E64d delicate) in TMPRSS2-negative cells akin to Hela-ACE2 when compared with earlier VOCs akin to Delta, while access into Calu-3 cells is in large part TMPRSS2 dependent (camostat delicate) (Prolonged Information Fig. 1a,b), leading to hanging mobile type-specific variations between variant titres by means of TCID50 (Prolonged Information Fig. 1e).Fig. 1: BA.5 shows enhanced innate immune antagonism throughout an infection of airway epithelial cells.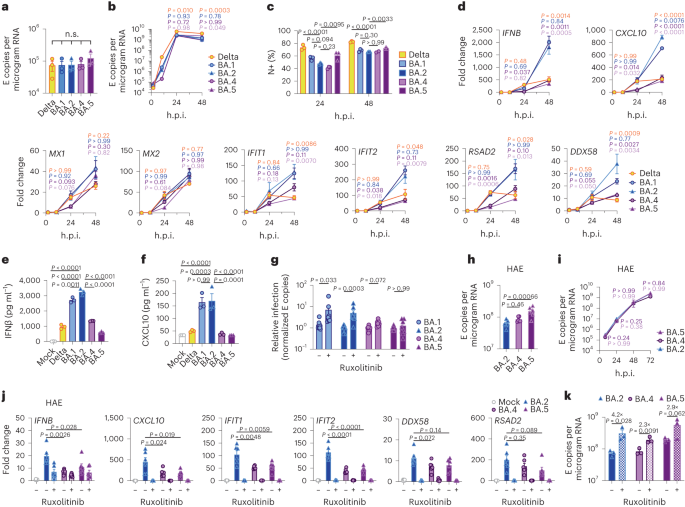 a–g, Calu-3 an infection with 2,000 E copies in keeping with mobile of Delta (yellow, Ο), BA.1 (blue, Ο), BA.2 (blue, Δ), BA.4 (pink, O) and BA.5 (pink, Δ), n = 3: imply viral E copies at 2 h.p.i. throughout 3 impartial experiments (a); viral replication over the years measured by means of RT–qPCR for intracellular E copies in keeping with microgram RNA (b); an infection ranges measured by means of nucleocapsid expression (% N+ by means of glide cytometry) (c); expression of IFNB, CXCL10, IFIT1, IFIT2, RSAD2, MX1, MX2 and DDX58 in contaminated cells over the years (d); IFNβ (e) and CXCL10 (f) secretion from contaminated Calu-3 cells measured by means of ELISA at 48 h.p.i.; rescue of viral replication by means of JAK1-inhibitor ruxolitinib in Calu-3 cells at 48 h.p.i., the place relative an infection ranges are proven throughout 3 impartial experiments made up our minds by means of E copies in keeping with microgram RNA normalized to the median an infection degree of the untreated keep an eye on (g). h–ok, Number one bronchial HAEs have been contaminated with the indicated variants at 1,500 E copies in keeping with mobile: viral replication measured by means of intracellular E copies at 72 h.p.i. (h) and viral unlock into apical washes over the years (i), with 3 organic replicates proven; expression of IFNB, CXCL10, IFIT1, IFIT2, DDX58 and RSAD2 in HAEs at 72 h.p.i., with six organic replicates proven (j); intracellular viral E copies in HAEs within the presence or absence of five μM ruxolitinib at 72 h.p.i., with 3 organic replicates proven (ok). For a, one-way research of variance (ANOVA) with Bonferroni post-test was once used. n.s., now not vital at P > 0.05 for all comparisons. For b–h and j, one-way ANOVA and Dunnett’s post-test have been used. For i, two-way ANOVA with a Bonferroni post-test was once used. For ok, one-tailed unpaired Pupil’s t-test was once used. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven. h.p.i., hours submit an infection.Supply dataInfection of Calu-3 cells with 2,000 E gene copies in keeping with mobile (Fig. 1) or 200 E copies in keeping with mobile (Prolonged Information Fig. 1) gave related E RNA (RT–qPCR) at 2 h submit an infection (h.p.i.), in line with equivalent enter doses (Fig. 1a and Prolonged Information Fig. 1g). E gene measurements throughout an infection printed that Omicron isolates BA.1, BA.2, BA.4 and BA.5 replicated in a similar fashion, lagging at the back of Delta in Calu-3 cells (Fig. 1b and Prolonged Information Fig. 1h–l). BA.4 replicated maximum slowly to begin with however stuck up with BA.1, BA.2 and BA.5 by means of 24 h.p.i. (Fig. 1b and Prolonged Information Fig. 1). Importantly, those replication variations have been seen constantly throughout a number of experiments (Fig. 1 and Prolonged Information Figs. 1 and a pair of). As E gene dimension throughout an infection captures genomic RNA (gRNA) in addition to E, S and Orf3 subgenomic RNAs (sgRNAs), we when compared the degrees of intracellular E RNA with the ones of Nsp12 and Orf1a (evaluate Prolonged Information Fig. 1h,i with Fig. 1b and Prolonged Information Fig. 1k,l with Prolonged Information Fig. 1j), which might be uniquely encoded inside gRNA. Importantly, the ratio of E to Nsp12 was once identical till 24 h.p.i. reflecting identical ranges of E sgRNA synthesis between variants (Prolonged Information Fig. 1m). Quantification of launched virions by means of measuring E and Nsp12 RNA copies within the supernatant reflected viral replication (Prolonged Information Fig. 1n–q). An identical patterns of an infection have been additionally noticed when quantified by means of intracellular nucleocapsid (N) staining (Fig. 1c and Prolonged Information Fig. 1r).BA.4 and BA.5 cause much less innate immune activation than earliest Omicron subvariantsWe subsequent when compared the host innate immune reaction to Omicron subvariant an infection of Calu-3 cells. All viral shares have been ready in human gastrointestinal Caco-2 cells as they’re naturally permissive to SARS-CoV-2 replication however don’t mount a powerful innate reaction to this infection19,26. We showed that viral shares ready in Caco-2 cells (the best viral inoculum for each and every variant was once 2,000 E copies in keeping with mobile) didn’t include measurable interferon (IFN)β and negligible IFNλ1/IFNλ3 (enzyme-linked immunosorbent assay, ELISA) (Prolonged Information Fig. 2a,b), making sure variations in innate immune activation in Calu-3 infections weren’t a results of IFN carryover within the viral shares.Strikingly, we discovered that an infection of Calu-3 cells with BA.4 and BA.5 ended in considerably much less innate immune activation in comparison to BA.1/BA.2, evidenced by means of decrease induction of IFNβ (IFNB) and interferon stimulated genes (ISGs) together with inflammatory chemokine CXCL10 and RSAD2, DDX58, IFIT1 and IFIT2 (Fig. 1d and Prolonged Information Fig. 2c–g) and a pattern in opposition to decreased MX1 and MX2 expression (Fig. 1d). Decreased host responses to BA.4 and BA.5 an infection have been additionally obtrusive on the degree of IFNβ and CXCL10 secretion (Fig. 1e,f). Slower replication of BA.4 most likely contributes partially to decreased innate immune activation throughout Calu-3 an infection, however BA.5 replication was once very similar to BA.1 and BA.2 and however prompted considerably much less innate immune responses. Inhibition of IFN-mediated JAK/STAT signalling with ruxolitinib, evidenced by means of the absence of ISG induction (Prolonged Information Fig. 2e,f), rescued BA.1 and BA.2 an infection in Calu-3 cells to a better level than BA.4 or BA.5 (Fig. 1g and Prolonged Information Fig. 2h–j), suggesting that the larger induction of IFNβ by means of BA.1 and BA.2 decreased their infectivity. BA.1 to BA.5 confirmed identical sensitivities to a variety of IFN doses used to pre-treat Calu-3 cells (Prolonged Information Fig. 2k–m). We subsequently conclude that the variations in ruxolitinib sensitivity mirror variations in IFN induction after Calu-3 an infection and now not variations in IFN sensitivity. Infecting Calu-3 cells with decrease virus enter doses (200 E copies in keeping with mobile) recapitulated our remark that Delta replicated higher than Omicron BA.1–BA.5 (Prolonged Information Fig. 1j–l), and we once more noticed decreased innate immune activation by means of BA.4 and BA.5 when compared with BA.1 and BA.2 (Prolonged Information Fig. 2f,g). At this decrease inoculum, BA.4 infectivity was once additionally strongly rescued by means of ruxolitinib remedy in line with its slower replication being because of IFN induction (Prolonged Information Fig. 2i).We subsequent when compared Omicron subvariant replication and host responses in number one HAE cultures, which higher recapitulate the heterogeneous polarized epithelial layer of the breathing tract. We have now up to now reported that HAEs disclose variations in VOC replication that most likely mirror host adaptation, which don’t seem to be all the time obvious in extremely permissive mobile strains, akin to Calu-3 (refs. 1,2). Concordantly, BA.5 viral replication was once upper than BA.2 and BA.4 in differentiated number one bronchial HAEs at 72 h.p.i., whilst apical viral unlock over the years was once related (Fig. 1h,i). Regardless of BA.4 and BA.5 replicating in a similar fashion to BA.2 in HAEs, we constantly seen decreased innate activation, measured by means of ISG induction, after BA.4 and BA.5 an infection (IFNB, CXCL10, IFIT1, IFIT2, DDX58 and RSAD2; Fig. 1j). Inhibiting IFN signalling with JAK-inhibitor ruxolitinib suppressed ISG induction (Fig. 1j) and rescued replication of BA.2 to a better level than BA.4 and BA.5 (Fig. 1k). Altogether, knowledge in Fig. 1 counsel adaptation to scale back innate immune activation between the earliest (BA.1 and BA.2) and next (BA.4 and BA.5) Omicron subvariants.SARS-CoV-2, and different breathing viruses, reportedly reflect extra successfully in nasal and tracheal epithelial cells27, partially because of decreased innate activation and IFN responsiveness on the decrease temperatures of the higher airway28,29,30. To analyze whether or not decrease temperatures disclose additional Omicron subvariant adaptation, we when compared replication at 32 °C in Calu-3 cells. We discovered BA.1 to BA.5 all replicated much less smartly than at 37 °C (Prolonged Information Fig. 3a,b) while Delta replication was once now not as temperature delicate. As expected29, innate immune activation in line with an infection, or to RNA sensing agonist poly(I:C), was once in large part abolished at 32 °C (measured by means of IFNB and CXCL10 messenger RNA induction; Prolonged Information Fig. 3c–e). At 37 °C, we once more seen decrease innate activation for BA.4 and BA.5 when compared with BA.1/BA.2. In HAE, reducing the temperature to 32 °C didn’t have an effect on viral replication to the similar extent as in Calu-3 cells (Prolonged Information Fig. 3f). Then again, we seen decreased virus output in apical washes from contaminated HAE cultures for all Omicron isolates (Prolonged Information Fig. 3g–i). Inflamed HAEs at 32 °C additionally expressed considerably much less IFNB and CXCL10 (Prolonged Information Fig. 3j). General, our knowledge counsel that Omicron does now not reflect higher at 32 °C in lung epithelial cells even within the absence of an innate immune reaction. Then again, it’s imaginable that the intra-tissue temperature all through the airlines stays nearer to 37 °C than the exhaled breath temperature of 32 °C suggests31.BA.4 and BA.5 build up Orf6 expression and successfully antagonize innate immune activation throughout infectionWe subsequent investigated the mechanism underlying differential innate immune activation by means of Omicron subvariants. IRF3 and STAT1 are key transcription components responding to intracellular RNA sensing and IFN manufacturing, respectively, exemplified right here by means of poly(I:C) remedy (Prolonged Information Fig. 4a–c). We and others have proven that SARS-CoV-2 turns on transcription components IRF3 and STAT1 downstream of RNA sensing19,32. In line with their decreased innate immune triggering, we discovered Omicron BA.4 and BA.5 an infection activated considerably much less IRF3 phosphorylation than BA.2 an infection (Fig. 2a–c). A identical pattern was once seen for STAT1 serine 727 phosphorylation, which is very important for complete STAT1 transcriptional activity33, however now not upstream JAK1-dependent tyrosine 701 phosphorylation (Fig. 2a,d–f). Aid of STAT1 phosphorylation correlated with decreased STAT1 nuclear translocation throughout BA.4 and BA.5 an infection when compared with BA.2, measured by means of high-content single-cell immunofluorescence imaging of contaminated nucleocapsid-positive Calu-3 cells (Fig. 2g). Those knowledge counsel that BA.4 and BA.5 extra successfully save you intracellular activation of innate sensing pathways. We up to now reported that SARS-CoV-2 VOC Alpha advanced enhanced innate immune evasion by means of expanding expression of key innate antagonists Orf6, Orf9b and N (Prolonged Information Fig. 4d), which manipulate host mobile innate immune pathways1. To analyze whether or not Omicron subvariants have additionally independently advanced enhanced innate immune suppression via identical mechanisms throughout human adaptation, we measured viral innate antagonist protein expression throughout an infection. Strikingly, we discovered that BA.4, and in particular BA.5, expressed upper ranges of Orf6 and N when compared with BA.1 and BA.2 (Fig. 2h–l and Prolonged Information Fig. 4e–ok), measured at 48 h.p.i. in Calu-3 cells when E RNA ranges have been identical (Fig. 2m). In contrast to earlier VOCs1,2, expression of innate immune antagonist Orf9b was once now not detected for any Omicron isolate, perhaps because of Omicron subvariants encoding lineage-specific Orf9b mutations (P10S and ΔENA at positions 27–29) changing antibody binding and precluding detection by means of immunoblot (Fig. 2n and Prolonged Information Fig. 4d). Importantly, Orf9b remained readily detectable in Delta-infected cells (Fig. 2n). Upregulation of Orf6 and N expression by means of BA.5 was once validated the use of a 2d impartial isolate (Prolonged Information Fig. 4l–n), and was once additionally obtrusive in lysates from contaminated HAEs (Prolonged Information Fig. 4o). Blockading IFN signalling with ruxolitinib rescued replication of all Omicron isolates as sooner than (Fig. 1 and Prolonged Information Fig. 2) and enhanced viral protein detection by means of immunoblot (Fig. 2h,n and Prolonged Information Fig. 4e). Importantly, upper ranges of BA.4 and BA.5 Orf6 and N remained obvious after ruxolitinib remedy (Fig. 2h,ok,l). We up to now confirmed that enhanced ranges of Orf6, N and Orf9b protein by means of Alpha have been related to greater ranges of the corresponding sgRNAs1. In contrast, BA.5 Orf6 and N sgRNA ranges (normalized to genomic Orf1a) weren’t enhanced, and have been most effective reasonably upregulated throughout BA.4 an infection (Fig. 2o), in particular compared to Alpha (Prolonged Information Fig. 4p,q). No variations have been seen in S and Orf3a sgRNAs, which served as controls to rule out a common enhancement of sgRNA synthesis (Fig. 2o). Even supposing Omicron subvariants have synonymous and non-synonymous mutations in Orf6 and N, there are not any mutations that distinguish BA.4 and BA.5 from BA.1 and BA.2 that offer a easy reason for greater Orf6 or N protein ranges, together with of their transcriptional regulatory sequences (Figs. 1 and a pair of and Prolonged Information Tables 1 and a pair of). Thus, we hypothesize that BA.4 and BA.5 have both advanced impartial mechanisms to extend Orf6 and N protein ranges, or that the rise is mediated by means of adjustments in different places within the genome, which would possibly have an effect on viral translation or protein balance. Additional research are required to pinpoint the diversifications regulating Orf6 and N expression ranges.Fig. 2: BA.5 successfully expresses SARS-CoV-2 innate antagonists throughout airway epithelial mobile an infection.
a–g, Calu-3 an infection with 2,000 E copies in keeping with mobile of Delta (yellow, Ο), BA.1 (blue, Ο), BA.2 (blue, Δ), BA.4 (pink, O) and BA.5 (pink, Δ), n = 3: imply viral E copies at 2 h.p.i. throughout 3 impartial experiments (a); viral replication over the years measured by means of RT–qPCR for intracellular E copies in keeping with microgram RNA (b); an infection ranges measured by means of nucleocapsid expression (% N+ by means of glide cytometry) (c); expression of IFNB, CXCL10, IFIT1, IFIT2, RSAD2, MX1, MX2 and DDX58 in contaminated cells over the years (d); IFNβ (e) and CXCL10 (f) secretion from contaminated Calu-3 cells measured by means of ELISA at 48 h.p.i.; rescue of viral replication by means of JAK1-inhibitor ruxolitinib in Calu-3 cells at 48 h.p.i., the place relative an infection ranges are proven throughout 3 impartial experiments made up our minds by means of E copies in keeping with microgram RNA normalized to the median an infection degree of the untreated keep an eye on (g). h–ok, Number one bronchial HAEs have been contaminated with the indicated variants at 1,500 E copies in keeping with mobile: viral replication measured by means of intracellular E copies at 72 h.p.i. (h) and viral unlock into apical washes over the years (i), with 3 organic replicates proven; expression of IFNB, CXCL10, IFIT1, IFIT2, DDX58 and RSAD2 in HAEs at 72 h.p.i., with six organic replicates proven (j); intracellular viral E copies in HAEs within the presence or absence of five μM ruxolitinib at 72 h.p.i., with 3 organic replicates proven (ok). For a, one-way research of variance (ANOVA) with Bonferroni post-test was once used. n.s., now not vital at P > 0.05 for all comparisons. For b–h and j, one-way ANOVA and Dunnett’s post-test have been used. For i, two-way ANOVA with a Bonferroni post-test was once used. For ok, one-tailed unpaired Pupil’s t-test was once used. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven. h.p.i., hours submit an infection.Supply dataInfection of Calu-3 cells with 2,000 E gene copies in keeping with mobile (Fig. 1) or 200 E copies in keeping with mobile (Prolonged Information Fig. 1) gave related E RNA (RT–qPCR) at 2 h submit an infection (h.p.i.), in line with equivalent enter doses (Fig. 1a and Prolonged Information Fig. 1g). E gene measurements throughout an infection printed that Omicron isolates BA.1, BA.2, BA.4 and BA.5 replicated in a similar fashion, lagging at the back of Delta in Calu-3 cells (Fig. 1b and Prolonged Information Fig. 1h–l). BA.4 replicated maximum slowly to begin with however stuck up with BA.1, BA.2 and BA.5 by means of 24 h.p.i. (Fig. 1b and Prolonged Information Fig. 1). Importantly, those replication variations have been seen constantly throughout a number of experiments (Fig. 1 and Prolonged Information Figs. 1 and a pair of). As E gene dimension throughout an infection captures genomic RNA (gRNA) in addition to E, S and Orf3 subgenomic RNAs (sgRNAs), we when compared the degrees of intracellular E RNA with the ones of Nsp12 and Orf1a (evaluate Prolonged Information Fig. 1h,i with Fig. 1b and Prolonged Information Fig. 1k,l with Prolonged Information Fig. 1j), which might be uniquely encoded inside gRNA. Importantly, the ratio of E to Nsp12 was once identical till 24 h.p.i. reflecting identical ranges of E sgRNA synthesis between variants (Prolonged Information Fig. 1m). Quantification of launched virions by means of measuring E and Nsp12 RNA copies within the supernatant reflected viral replication (Prolonged Information Fig. 1n–q). An identical patterns of an infection have been additionally noticed when quantified by means of intracellular nucleocapsid (N) staining (Fig. 1c and Prolonged Information Fig. 1r).BA.4 and BA.5 cause much less innate immune activation than earliest Omicron subvariantsWe subsequent when compared the host innate immune reaction to Omicron subvariant an infection of Calu-3 cells. All viral shares have been ready in human gastrointestinal Caco-2 cells as they’re naturally permissive to SARS-CoV-2 replication however don’t mount a powerful innate reaction to this infection19,26. We showed that viral shares ready in Caco-2 cells (the best viral inoculum for each and every variant was once 2,000 E copies in keeping with mobile) didn’t include measurable interferon (IFN)β and negligible IFNλ1/IFNλ3 (enzyme-linked immunosorbent assay, ELISA) (Prolonged Information Fig. 2a,b), making sure variations in innate immune activation in Calu-3 infections weren’t a results of IFN carryover within the viral shares.Strikingly, we discovered that an infection of Calu-3 cells with BA.4 and BA.5 ended in considerably much less innate immune activation in comparison to BA.1/BA.2, evidenced by means of decrease induction of IFNβ (IFNB) and interferon stimulated genes (ISGs) together with inflammatory chemokine CXCL10 and RSAD2, DDX58, IFIT1 and IFIT2 (Fig. 1d and Prolonged Information Fig. 2c–g) and a pattern in opposition to decreased MX1 and MX2 expression (Fig. 1d). Decreased host responses to BA.4 and BA.5 an infection have been additionally obtrusive on the degree of IFNβ and CXCL10 secretion (Fig. 1e,f). Slower replication of BA.4 most likely contributes partially to decreased innate immune activation throughout Calu-3 an infection, however BA.5 replication was once very similar to BA.1 and BA.2 and however prompted considerably much less innate immune responses. Inhibition of IFN-mediated JAK/STAT signalling with ruxolitinib, evidenced by means of the absence of ISG induction (Prolonged Information Fig. 2e,f), rescued BA.1 and BA.2 an infection in Calu-3 cells to a better level than BA.4 or BA.5 (Fig. 1g and Prolonged Information Fig. 2h–j), suggesting that the larger induction of IFNβ by means of BA.1 and BA.2 decreased their infectivity. BA.1 to BA.5 confirmed identical sensitivities to a variety of IFN doses used to pre-treat Calu-3 cells (Prolonged Information Fig. 2k–m). We subsequently conclude that the variations in ruxolitinib sensitivity mirror variations in IFN induction after Calu-3 an infection and now not variations in IFN sensitivity. Infecting Calu-3 cells with decrease virus enter doses (200 E copies in keeping with mobile) recapitulated our remark that Delta replicated higher than Omicron BA.1–BA.5 (Prolonged Information Fig. 1j–l), and we once more noticed decreased innate immune activation by means of BA.4 and BA.5 when compared with BA.1 and BA.2 (Prolonged Information Fig. 2f,g). At this decrease inoculum, BA.4 infectivity was once additionally strongly rescued by means of ruxolitinib remedy in line with its slower replication being because of IFN induction (Prolonged Information Fig. 2i).We subsequent when compared Omicron subvariant replication and host responses in number one HAE cultures, which higher recapitulate the heterogeneous polarized epithelial layer of the breathing tract. We have now up to now reported that HAEs disclose variations in VOC replication that most likely mirror host adaptation, which don’t seem to be all the time obvious in extremely permissive mobile strains, akin to Calu-3 (refs. 1,2). Concordantly, BA.5 viral replication was once upper than BA.2 and BA.4 in differentiated number one bronchial HAEs at 72 h.p.i., whilst apical viral unlock over the years was once related (Fig. 1h,i). Regardless of BA.4 and BA.5 replicating in a similar fashion to BA.2 in HAEs, we constantly seen decreased innate activation, measured by means of ISG induction, after BA.4 and BA.5 an infection (IFNB, CXCL10, IFIT1, IFIT2, DDX58 and RSAD2; Fig. 1j). Inhibiting IFN signalling with JAK-inhibitor ruxolitinib suppressed ISG induction (Fig. 1j) and rescued replication of BA.2 to a better level than BA.4 and BA.5 (Fig. 1k). Altogether, knowledge in Fig. 1 counsel adaptation to scale back innate immune activation between the earliest (BA.1 and BA.2) and next (BA.4 and BA.5) Omicron subvariants.SARS-CoV-2, and different breathing viruses, reportedly reflect extra successfully in nasal and tracheal epithelial cells27, partially because of decreased innate activation and IFN responsiveness on the decrease temperatures of the higher airway28,29,30. To analyze whether or not decrease temperatures disclose additional Omicron subvariant adaptation, we when compared replication at 32 °C in Calu-3 cells. We discovered BA.1 to BA.5 all replicated much less smartly than at 37 °C (Prolonged Information Fig. 3a,b) while Delta replication was once now not as temperature delicate. As expected29, innate immune activation in line with an infection, or to RNA sensing agonist poly(I:C), was once in large part abolished at 32 °C (measured by means of IFNB and CXCL10 messenger RNA induction; Prolonged Information Fig. 3c–e). At 37 °C, we once more seen decrease innate activation for BA.4 and BA.5 when compared with BA.1/BA.2. In HAE, reducing the temperature to 32 °C didn’t have an effect on viral replication to the similar extent as in Calu-3 cells (Prolonged Information Fig. 3f). Then again, we seen decreased virus output in apical washes from contaminated HAE cultures for all Omicron isolates (Prolonged Information Fig. 3g–i). Inflamed HAEs at 32 °C additionally expressed considerably much less IFNB and CXCL10 (Prolonged Information Fig. 3j). General, our knowledge counsel that Omicron does now not reflect higher at 32 °C in lung epithelial cells even within the absence of an innate immune reaction. Then again, it’s imaginable that the intra-tissue temperature all through the airlines stays nearer to 37 °C than the exhaled breath temperature of 32 °C suggests31.BA.4 and BA.5 build up Orf6 expression and successfully antagonize innate immune activation throughout infectionWe subsequent investigated the mechanism underlying differential innate immune activation by means of Omicron subvariants. IRF3 and STAT1 are key transcription components responding to intracellular RNA sensing and IFN manufacturing, respectively, exemplified right here by means of poly(I:C) remedy (Prolonged Information Fig. 4a–c). We and others have proven that SARS-CoV-2 turns on transcription components IRF3 and STAT1 downstream of RNA sensing19,32. In line with their decreased innate immune triggering, we discovered Omicron BA.4 and BA.5 an infection activated considerably much less IRF3 phosphorylation than BA.2 an infection (Fig. 2a–c). A identical pattern was once seen for STAT1 serine 727 phosphorylation, which is very important for complete STAT1 transcriptional activity33, however now not upstream JAK1-dependent tyrosine 701 phosphorylation (Fig. 2a,d–f). Aid of STAT1 phosphorylation correlated with decreased STAT1 nuclear translocation throughout BA.4 and BA.5 an infection when compared with BA.2, measured by means of high-content single-cell immunofluorescence imaging of contaminated nucleocapsid-positive Calu-3 cells (Fig. 2g). Those knowledge counsel that BA.4 and BA.5 extra successfully save you intracellular activation of innate sensing pathways. We up to now reported that SARS-CoV-2 VOC Alpha advanced enhanced innate immune evasion by means of expanding expression of key innate antagonists Orf6, Orf9b and N (Prolonged Information Fig. 4d), which manipulate host mobile innate immune pathways1. To analyze whether or not Omicron subvariants have additionally independently advanced enhanced innate immune suppression via identical mechanisms throughout human adaptation, we measured viral innate antagonist protein expression throughout an infection. Strikingly, we discovered that BA.4, and in particular BA.5, expressed upper ranges of Orf6 and N when compared with BA.1 and BA.2 (Fig. 2h–l and Prolonged Information Fig. 4e–ok), measured at 48 h.p.i. in Calu-3 cells when E RNA ranges have been identical (Fig. 2m). In contrast to earlier VOCs1,2, expression of innate immune antagonist Orf9b was once now not detected for any Omicron isolate, perhaps because of Omicron subvariants encoding lineage-specific Orf9b mutations (P10S and ΔENA at positions 27–29) changing antibody binding and precluding detection by means of immunoblot (Fig. 2n and Prolonged Information Fig. 4d). Importantly, Orf9b remained readily detectable in Delta-infected cells (Fig. 2n). Upregulation of Orf6 and N expression by means of BA.5 was once validated the use of a 2d impartial isolate (Prolonged Information Fig. 4l–n), and was once additionally obtrusive in lysates from contaminated HAEs (Prolonged Information Fig. 4o). Blockading IFN signalling with ruxolitinib rescued replication of all Omicron isolates as sooner than (Fig. 1 and Prolonged Information Fig. 2) and enhanced viral protein detection by means of immunoblot (Fig. 2h,n and Prolonged Information Fig. 4e). Importantly, upper ranges of BA.4 and BA.5 Orf6 and N remained obvious after ruxolitinib remedy (Fig. 2h,ok,l). We up to now confirmed that enhanced ranges of Orf6, N and Orf9b protein by means of Alpha have been related to greater ranges of the corresponding sgRNAs1. In contrast, BA.5 Orf6 and N sgRNA ranges (normalized to genomic Orf1a) weren’t enhanced, and have been most effective reasonably upregulated throughout BA.4 an infection (Fig. 2o), in particular compared to Alpha (Prolonged Information Fig. 4p,q). No variations have been seen in S and Orf3a sgRNAs, which served as controls to rule out a common enhancement of sgRNA synthesis (Fig. 2o). Even supposing Omicron subvariants have synonymous and non-synonymous mutations in Orf6 and N, there are not any mutations that distinguish BA.4 and BA.5 from BA.1 and BA.2 that offer a easy reason for greater Orf6 or N protein ranges, together with of their transcriptional regulatory sequences (Figs. 1 and a pair of and Prolonged Information Tables 1 and a pair of). Thus, we hypothesize that BA.4 and BA.5 have both advanced impartial mechanisms to extend Orf6 and N protein ranges, or that the rise is mediated by means of adjustments in different places within the genome, which would possibly have an effect on viral translation or protein balance. Additional research are required to pinpoint the diversifications regulating Orf6 and N expression ranges.Fig. 2: BA.5 successfully expresses SARS-CoV-2 innate antagonists throughout airway epithelial mobile an infection.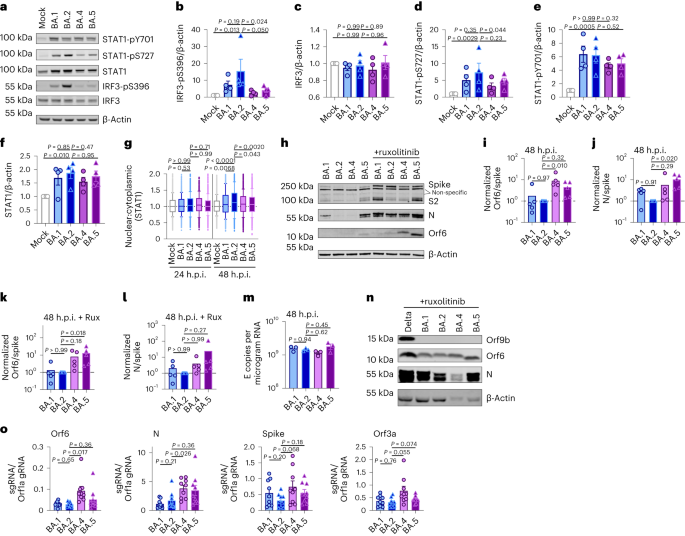 Calu-3 cells have been contaminated with 2,000 E copies in keeping with mobile of the indicated variants. a, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin at 24 h.p.i. One among 4 impartial western blots is proven. b–f, Quantification of 4 impartial western blots appearing IRF3-pS396 (b), IRF3 (c), STAT1-pS727 (d), STAT1-pY701 (e) and STAT1 (f) over β-actin at 24 h.p.i. normalized to mock. g, Quantification of STAT1 nuclear translocation detected by means of single-cell fluorescence microscopy over the years in Calu-3 cells contaminated with the indicated variants. Information from 1,500 cells in keeping with situation are proven. In contaminated cultures, translocation was once made up our minds in N+ cells. h, Western blot of Orf6, N, spike and β-actin at 48 h.p.i. in contaminated cells ± 5 μM ruxolitinib (Rux). Non-specific bands detected by means of polyclonal anti-spike number one antibody are indicated (see Prolonged Information Fig. 4e for mock). One among 5 impartial western blots proven. i–l, Quantification of Orf6 and N expression from 5 impartial western blots of Calu-3 cells within the absence (i, Orf6; j, N) or presence of five μM ruxolitinib (ok, Orf6; l, N) at 48 h.p.i., normalized to spike over BA.2. m, Viral replication in cells from h. n, Consultant western blot of Calu-3 cells contaminated with Delta, BA.1, BA.2, BA.4 and BA.5 at 2,000 E copies in keeping with mobile appearing Orf9b, Orf6, N and β-actin expression at 48 h.p.i. + 5 μM ruxolitinib. o, sgRNA expression of Orf6, N, spike and Orf3a normalized to Orf1a gRNA in Calu-3 cells at 48 h.p.i.; 9 measurements from 3 impartial experiments proven. For b–f, i–m and o, one-way research of variance with Dunnett’s post-test was once used. For g, box-and-whisker blots display tenth–ninetieth percentile, and teams have been when compared at each and every timepoint as indicated the use of a Kruskal–Wallis check. Imply ± s.e.m. or particular person datapoints are proven.Supply dataOrf6 expression is a significant determinant of enhanced innate immune antagonism by means of rising VOCsOrf6 is a multifunctional viral accent protein that modulates expression of host and viral proteins34,35. Orf6 selectively inhibits host transcription issue nuclear delivery to potently antagonize antiviral responses throughout an infection. To probe Orf6 mechanisms, and its contribution to enhanced innate antagonism by means of the VOCs, we used opposite genetics to introduce two prevent codons into the Orf6 coding collection of each Alpha (Alpha ΔOrf6) and BA.5 (BA.5 ΔOrf6), which we showed abolished Orf6 expression throughout an infection (Fig. 3a,b). Whilst Alpha ΔOrf6 replicated in a similar fashion to parental wild-type (WT) virus as much as 24 h.p.i. (Fig. 3c), we seen enhanced IFNB and CXCL10 expression (Fig. 3d) and protein secretion (Prolonged Information Fig. 5a) throughout Alpha ΔOrf6 an infection of Calu-3 cells when compared with WT virus. Additionally, greater IRF3 nuclear translocation was once obtrusive after Alpha ΔOrf6 an infection at 24 h.p.i. the use of single-cell quantitative immunofluorescence microscopy (Fig. 3e and Prolonged Information Fig. 5b). This implies a very powerful function for Orf6 in innate immune antagonism throughout viral replication1,35,36,37 and is in line with suppression of IRF3 nuclear delivery in Orf6 overexpression studies35,36,38. The aid in Alpha ΔOrf6 replication at 48 h.p.i., and N and spike protein expression at 24 h.p.i., that was once rescued by means of ruxolitinib remedy, may be in line with larger IFN-mediated suppression of the Orf6 deletion mutant (Fig. 3a and Prolonged Information Fig. 5c).Fig. 3: Orf6 expression is a significant determinant of enhanced innate immune antagonism by means of rising VOCs.
Calu-3 cells have been contaminated with 2,000 E copies in keeping with mobile of the indicated variants. a, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin at 24 h.p.i. One among 4 impartial western blots is proven. b–f, Quantification of 4 impartial western blots appearing IRF3-pS396 (b), IRF3 (c), STAT1-pS727 (d), STAT1-pY701 (e) and STAT1 (f) over β-actin at 24 h.p.i. normalized to mock. g, Quantification of STAT1 nuclear translocation detected by means of single-cell fluorescence microscopy over the years in Calu-3 cells contaminated with the indicated variants. Information from 1,500 cells in keeping with situation are proven. In contaminated cultures, translocation was once made up our minds in N+ cells. h, Western blot of Orf6, N, spike and β-actin at 48 h.p.i. in contaminated cells ± 5 μM ruxolitinib (Rux). Non-specific bands detected by means of polyclonal anti-spike number one antibody are indicated (see Prolonged Information Fig. 4e for mock). One among 5 impartial western blots proven. i–l, Quantification of Orf6 and N expression from 5 impartial western blots of Calu-3 cells within the absence (i, Orf6; j, N) or presence of five μM ruxolitinib (ok, Orf6; l, N) at 48 h.p.i., normalized to spike over BA.2. m, Viral replication in cells from h. n, Consultant western blot of Calu-3 cells contaminated with Delta, BA.1, BA.2, BA.4 and BA.5 at 2,000 E copies in keeping with mobile appearing Orf9b, Orf6, N and β-actin expression at 48 h.p.i. + 5 μM ruxolitinib. o, sgRNA expression of Orf6, N, spike and Orf3a normalized to Orf1a gRNA in Calu-3 cells at 48 h.p.i.; 9 measurements from 3 impartial experiments proven. For b–f, i–m and o, one-way research of variance with Dunnett’s post-test was once used. For g, box-and-whisker blots display tenth–ninetieth percentile, and teams have been when compared at each and every timepoint as indicated the use of a Kruskal–Wallis check. Imply ± s.e.m. or particular person datapoints are proven.Supply dataOrf6 expression is a significant determinant of enhanced innate immune antagonism by means of rising VOCsOrf6 is a multifunctional viral accent protein that modulates expression of host and viral proteins34,35. Orf6 selectively inhibits host transcription issue nuclear delivery to potently antagonize antiviral responses throughout an infection. To probe Orf6 mechanisms, and its contribution to enhanced innate antagonism by means of the VOCs, we used opposite genetics to introduce two prevent codons into the Orf6 coding collection of each Alpha (Alpha ΔOrf6) and BA.5 (BA.5 ΔOrf6), which we showed abolished Orf6 expression throughout an infection (Fig. 3a,b). Whilst Alpha ΔOrf6 replicated in a similar fashion to parental wild-type (WT) virus as much as 24 h.p.i. (Fig. 3c), we seen enhanced IFNB and CXCL10 expression (Fig. 3d) and protein secretion (Prolonged Information Fig. 5a) throughout Alpha ΔOrf6 an infection of Calu-3 cells when compared with WT virus. Additionally, greater IRF3 nuclear translocation was once obtrusive after Alpha ΔOrf6 an infection at 24 h.p.i. the use of single-cell quantitative immunofluorescence microscopy (Fig. 3e and Prolonged Information Fig. 5b). This implies a very powerful function for Orf6 in innate immune antagonism throughout viral replication1,35,36,37 and is in line with suppression of IRF3 nuclear delivery in Orf6 overexpression studies35,36,38. The aid in Alpha ΔOrf6 replication at 48 h.p.i., and N and spike protein expression at 24 h.p.i., that was once rescued by means of ruxolitinib remedy, may be in line with larger IFN-mediated suppression of the Orf6 deletion mutant (Fig. 3a and Prolonged Information Fig. 5c).Fig. 3: Orf6 expression is a significant determinant of enhanced innate immune antagonism by means of rising VOCs.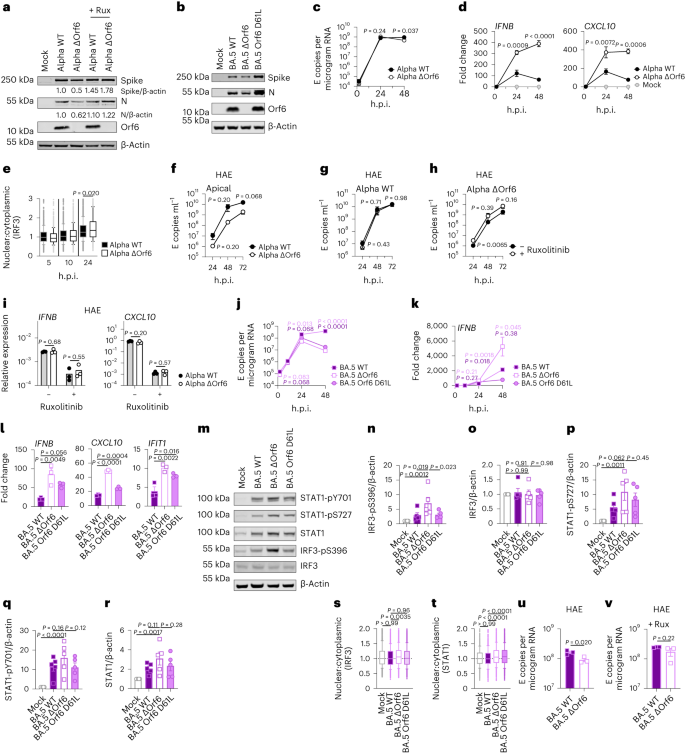 a,b, Western blot of Alpha (a) or BA.5 (b) opposite genetic (RG) virus infections in Calu-3 cells at 24 h.p.i. ± 5 μM ruxolitinib (Rux). c, Replication of RG viruses parental Alpha WT and ΔOrf6 in Calu-3 cells contaminated with 2,000 E copies in keeping with mobile over the years. d, Gene expression in cells from c over the years. e, Quantification of IRF3 nuclear translocation detected by means of single-cell fluorescence microscopy over the years. f–i, HAEs have been contaminated with 1,500 E copies in keeping with mobile of the indicated variants ± 5 μM ruxolitinib. f, Viral unlock into apical washes over the years. g,h, Apical unlock in HAEs contaminated with Alpha WT (g) or ΔOrf6 ± 5 μM ruxolitinib (h). i, Gene expression in cells from f. 3 organic replicates proven. j, Replication of RG viruses BA.5 WT, ΔOrf6 and Orf6 D61L isolates in Calu-3 cells contaminated with 2,000 E copies in keeping with mobile over the years. ok, IFNB expression in cells from j. l, Gene expression of Calu-3 cells at 24 h.p.i. m, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin at 24 h.p.i. n–r, Quantification of 5 impartial western blots appearing IRF3-pS396 (n), general IRF3 (o), STAT1-pS727 (p), STAT1-pY701 (q) and general STAT1 (r) over β-actin at 24 h.p.i. s,t, Quantification of IRF3 (s) and STAT1 (t) nuclear translocation detected by means of single-cell fluorescence microscopy at 24 h.p.i. u,v, Replication of BA.5 WT and ΔOrf6 in HAEs contaminated with 1,500 E copies in keeping with mobile within the absence (u) or presence (v) of five μM ruxolitinib. For c and d, two-way research of variance (ANOVA) and Bonferroni post-test have been used. For e, s and t, knowledge from 1,500 cells in keeping with situation are proven as box-and-whisker blots indicating tenth–ninetieth percentile. In contaminated cultures, translocation was once made up our minds in N+ cells. Teams have been when compared by means of Kruskal–Wallis check. For ok, l and n–r, one-way ANOVA with Dunnett’s post-test was once used. For f–i, u and v, unpaired two-tailed Pupil’s t-test was once used. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven.Supply dataAlpha ΔOrf6 additionally replicated much less smartly than WT in HAE cells (Fig. 3f–h and Prolonged Information Fig. 5d). IFNB and CXCL10 gene induction, normalized to GAPDH, have been identical after Alpha ΔOrf6 and WT an infection (Fig. 3i), regardless of decrease E RNA ranges for Alpha ΔOrf6, in line with greater innate immune induction by means of the deletion virus. Importantly, Alpha ΔOrf6 was once extra delicate to ruxolitinib remedy than WT, in line with the perception that greater IFN induction led to decreased replication of Alpha ΔOrf6 (Fig. 3g,h). To deal with the function of Orf6 throughout BA.5 an infection, we when compared replication of a BA.5 ΔOrf6 mutant with parental BA.5 WT virus. We additionally generated a BA.5 mutant bearing the Orf6 D61L mutation present in BA.2 and BA.4 that has been proposed to scale back Orf6 function2,32,39 (Fig. 3b,j). In line with the SARS-CoV-2 Alpha ΔOrf6 effects, BA.5 ΔOrf6 confirmed a replication defect at 48 h.p.i. when compared with BA.5 WT, and brought about considerably enhanced innate immune responses evidenced by means of enhanced IFNB and ISG induction (Fig. 3k,l). Deletion of Orf6 in BA.5 additionally greater the level of infection-induced IRF3 and STAT1 phosphorylation (Fig. 3m–r) and nuclear translocation (Fig. 3s,t). This demonstrates that Orf6 loss complements IRF3 and STAT1 activation regardless of identical ranges of an infection within the first 24 h.p.i., confirming the essential function of Orf6 in innate immune suppression and in distinguishing BA.5 from previous Omicron subvariants. An infection of HAEs showed decreased viral replication of BA.5 ΔOrf6 when compared with WT BA.5, whilst viral unlock remained related (Fig. 3u,v and Prolonged Information Fig. 5e). ISG expression in HAEs was once identical between WT and mutant regardless of decrease E RNA ranges throughout BA.5 ΔOrf6 an infection, suggesting larger induction of innate immunity within the absence of Orf6 in those cells (Prolonged Information Fig. 5f). Apparently, introducing the C-terminal D61L mutation into BA.5 Orf6 ended in an intermediate innate immune phenotype measured by means of greater induction of IFNB, CXCL10 and IFIT1 expression by means of the mutant virus (Fig. 3l). IRF3 phosphorylation and nuclear translocation have been identical between BA.5 WT and Orf6 D61L (Fig. 3n–s), while STAT1 translocation was once now not antagonized by means of Orf6 D61L (Fig. 3t), in keeping with reviews of a partial lack of Orf6 serve as within the D61L mutation2,32,39. Those knowledge counsel complicated adaptation of Orf6 manipulation of innate immunity throughout SARS-CoV-2 Omicron lineage adaptation.Enhanced innate antagonism is a conserved characteristic of dominant Omicron subvariantsDuring the process this find out about, SARS-CoV-2 has persisted to adapt and convey new Omicron subvariants (Fig. 4a and Prolonged Information Fig. 6a). Omicron subvariants BA.2.75, XBB.1, XBB.1.5 and BQ.1.1 have received greater ACE2 binding and enhanced adaptive immune evasion40,41,42,43. To check whether or not enhanced innate immune antagonism is continually related to globally a hit subvariants, we when compared BA.2.75, XBB.1, XBB.1.5 and BQ.1.1 isolates with BA.2 and BA.5 (Fig. 4). We equalized virus dose by means of Nsp12 RNA copies (RT–qPCR), a dimension of gRNA, quite than E RNA copies, because of accumulation of mutations within the E gene of later Omicron subvariants, together with within the area detected by means of our RT–qPCR assay. We discovered that each one Omicron subvariants retained an enhanced dependence on cathepsin, right here measured in A549 cells expressing ACE2 and TMPRSS2 (Prolonged Information Fig. 6b). BA.2.75, XBB.1 (two impartial isolates) and XBB.1.5, derived from the parental BA.2 lineage41,43, replicated comparably to previous BA.2 and BA.5 in Calu-3 and HAEs (Fig. 4b–e and Prolonged Information Fig. 6c–h). BQ.1.1, which has arisen from BA.5 (ref. 43), displayed some aid of replication in epithelial cells (Fig. 4d,e and Prolonged Information Fig. 6e,h). Very similar to BA.5, we discovered that each one next Omicron subvariants examined brought about considerably much less IFNB and CXCL10 expression than BA.2 at 24 h.p.i. (Fig. 4f). All Omicron subvariants derived from BA.2 (BA.2.75, XBB.1 and XBB.1.5) confirmed decreased rescue by means of ruxolitinib remedy, in addition to decreased induction of, or sensitivity to, IFN, very similar to BA.5 (Fig. 4g and Prolonged Information Fig. 6i). Strikingly, like BA.5, enhanced innate immune evasion by means of those newer subvariants was once accompanied by means of greater Orf6 expression for almost all of isolates (Fig. 4h,i). Decreased BQ.1.1 replication in Calu-3 cells (Fig. 4d and Prolonged Information Fig. 6e) avoided Orf6 and N detection within the absence of ruxolitinib (Fig. 4h). Decreased innate activation by means of fresh Omicron subvariants additionally correlated with decreased IRF3 phosphorylation when compared with BA.2, and aid of STAT1 serine phosphorylation was once mainly seen for XBB.1 and XBB.1.5 variants (Fig. 4j–l and Prolonged Information Fig. 6j–l). In combination those knowledge are in line with a pattern for ongoing Omicron evolution bettering Orf6 expression because it adapts to the human inhabitants resulting in decreased innate immune responses, detectable on the degree of IFN and ISG expression, and on the degree of transcription issue phosphorylation and nuclear translocation. This find out about bearing in mind Omicron variants may be very harking back to our earlier remark of enhanced expression of key innate immune antagonists Orf6, N and Orf9b in VOCs Alpha to Delta suggesting a not unusual evolutionary trajectory to combatting human innate immunity to beef up transmission1,2.Fig. 4: Innate immune phenotype of dominant Omicron subvariants.
a,b, Western blot of Alpha (a) or BA.5 (b) opposite genetic (RG) virus infections in Calu-3 cells at 24 h.p.i. ± 5 μM ruxolitinib (Rux). c, Replication of RG viruses parental Alpha WT and ΔOrf6 in Calu-3 cells contaminated with 2,000 E copies in keeping with mobile over the years. d, Gene expression in cells from c over the years. e, Quantification of IRF3 nuclear translocation detected by means of single-cell fluorescence microscopy over the years. f–i, HAEs have been contaminated with 1,500 E copies in keeping with mobile of the indicated variants ± 5 μM ruxolitinib. f, Viral unlock into apical washes over the years. g,h, Apical unlock in HAEs contaminated with Alpha WT (g) or ΔOrf6 ± 5 μM ruxolitinib (h). i, Gene expression in cells from f. 3 organic replicates proven. j, Replication of RG viruses BA.5 WT, ΔOrf6 and Orf6 D61L isolates in Calu-3 cells contaminated with 2,000 E copies in keeping with mobile over the years. ok, IFNB expression in cells from j. l, Gene expression of Calu-3 cells at 24 h.p.i. m, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin at 24 h.p.i. n–r, Quantification of 5 impartial western blots appearing IRF3-pS396 (n), general IRF3 (o), STAT1-pS727 (p), STAT1-pY701 (q) and general STAT1 (r) over β-actin at 24 h.p.i. s,t, Quantification of IRF3 (s) and STAT1 (t) nuclear translocation detected by means of single-cell fluorescence microscopy at 24 h.p.i. u,v, Replication of BA.5 WT and ΔOrf6 in HAEs contaminated with 1,500 E copies in keeping with mobile within the absence (u) or presence (v) of five μM ruxolitinib. For c and d, two-way research of variance (ANOVA) and Bonferroni post-test have been used. For e, s and t, knowledge from 1,500 cells in keeping with situation are proven as box-and-whisker blots indicating tenth–ninetieth percentile. In contaminated cultures, translocation was once made up our minds in N+ cells. Teams have been when compared by means of Kruskal–Wallis check. For ok, l and n–r, one-way ANOVA with Dunnett’s post-test was once used. For f–i, u and v, unpaired two-tailed Pupil’s t-test was once used. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven.Supply dataAlpha ΔOrf6 additionally replicated much less smartly than WT in HAE cells (Fig. 3f–h and Prolonged Information Fig. 5d). IFNB and CXCL10 gene induction, normalized to GAPDH, have been identical after Alpha ΔOrf6 and WT an infection (Fig. 3i), regardless of decrease E RNA ranges for Alpha ΔOrf6, in line with greater innate immune induction by means of the deletion virus. Importantly, Alpha ΔOrf6 was once extra delicate to ruxolitinib remedy than WT, in line with the perception that greater IFN induction led to decreased replication of Alpha ΔOrf6 (Fig. 3g,h). To deal with the function of Orf6 throughout BA.5 an infection, we when compared replication of a BA.5 ΔOrf6 mutant with parental BA.5 WT virus. We additionally generated a BA.5 mutant bearing the Orf6 D61L mutation present in BA.2 and BA.4 that has been proposed to scale back Orf6 function2,32,39 (Fig. 3b,j). In line with the SARS-CoV-2 Alpha ΔOrf6 effects, BA.5 ΔOrf6 confirmed a replication defect at 48 h.p.i. when compared with BA.5 WT, and brought about considerably enhanced innate immune responses evidenced by means of enhanced IFNB and ISG induction (Fig. 3k,l). Deletion of Orf6 in BA.5 additionally greater the level of infection-induced IRF3 and STAT1 phosphorylation (Fig. 3m–r) and nuclear translocation (Fig. 3s,t). This demonstrates that Orf6 loss complements IRF3 and STAT1 activation regardless of identical ranges of an infection within the first 24 h.p.i., confirming the essential function of Orf6 in innate immune suppression and in distinguishing BA.5 from previous Omicron subvariants. An infection of HAEs showed decreased viral replication of BA.5 ΔOrf6 when compared with WT BA.5, whilst viral unlock remained related (Fig. 3u,v and Prolonged Information Fig. 5e). ISG expression in HAEs was once identical between WT and mutant regardless of decrease E RNA ranges throughout BA.5 ΔOrf6 an infection, suggesting larger induction of innate immunity within the absence of Orf6 in those cells (Prolonged Information Fig. 5f). Apparently, introducing the C-terminal D61L mutation into BA.5 Orf6 ended in an intermediate innate immune phenotype measured by means of greater induction of IFNB, CXCL10 and IFIT1 expression by means of the mutant virus (Fig. 3l). IRF3 phosphorylation and nuclear translocation have been identical between BA.5 WT and Orf6 D61L (Fig. 3n–s), while STAT1 translocation was once now not antagonized by means of Orf6 D61L (Fig. 3t), in keeping with reviews of a partial lack of Orf6 serve as within the D61L mutation2,32,39. Those knowledge counsel complicated adaptation of Orf6 manipulation of innate immunity throughout SARS-CoV-2 Omicron lineage adaptation.Enhanced innate antagonism is a conserved characteristic of dominant Omicron subvariantsDuring the process this find out about, SARS-CoV-2 has persisted to adapt and convey new Omicron subvariants (Fig. 4a and Prolonged Information Fig. 6a). Omicron subvariants BA.2.75, XBB.1, XBB.1.5 and BQ.1.1 have received greater ACE2 binding and enhanced adaptive immune evasion40,41,42,43. To check whether or not enhanced innate immune antagonism is continually related to globally a hit subvariants, we when compared BA.2.75, XBB.1, XBB.1.5 and BQ.1.1 isolates with BA.2 and BA.5 (Fig. 4). We equalized virus dose by means of Nsp12 RNA copies (RT–qPCR), a dimension of gRNA, quite than E RNA copies, because of accumulation of mutations within the E gene of later Omicron subvariants, together with within the area detected by means of our RT–qPCR assay. We discovered that each one Omicron subvariants retained an enhanced dependence on cathepsin, right here measured in A549 cells expressing ACE2 and TMPRSS2 (Prolonged Information Fig. 6b). BA.2.75, XBB.1 (two impartial isolates) and XBB.1.5, derived from the parental BA.2 lineage41,43, replicated comparably to previous BA.2 and BA.5 in Calu-3 and HAEs (Fig. 4b–e and Prolonged Information Fig. 6c–h). BQ.1.1, which has arisen from BA.5 (ref. 43), displayed some aid of replication in epithelial cells (Fig. 4d,e and Prolonged Information Fig. 6e,h). Very similar to BA.5, we discovered that each one next Omicron subvariants examined brought about considerably much less IFNB and CXCL10 expression than BA.2 at 24 h.p.i. (Fig. 4f). All Omicron subvariants derived from BA.2 (BA.2.75, XBB.1 and XBB.1.5) confirmed decreased rescue by means of ruxolitinib remedy, in addition to decreased induction of, or sensitivity to, IFN, very similar to BA.5 (Fig. 4g and Prolonged Information Fig. 6i). Strikingly, like BA.5, enhanced innate immune evasion by means of those newer subvariants was once accompanied by means of greater Orf6 expression for almost all of isolates (Fig. 4h,i). Decreased BQ.1.1 replication in Calu-3 cells (Fig. 4d and Prolonged Information Fig. 6e) avoided Orf6 and N detection within the absence of ruxolitinib (Fig. 4h). Decreased innate activation by means of fresh Omicron subvariants additionally correlated with decreased IRF3 phosphorylation when compared with BA.2, and aid of STAT1 serine phosphorylation was once mainly seen for XBB.1 and XBB.1.5 variants (Fig. 4j–l and Prolonged Information Fig. 6j–l). In combination those knowledge are in line with a pattern for ongoing Omicron evolution bettering Orf6 expression because it adapts to the human inhabitants resulting in decreased innate immune responses, detectable on the degree of IFN and ISG expression, and on the degree of transcription issue phosphorylation and nuclear translocation. This find out about bearing in mind Omicron variants may be very harking back to our earlier remark of enhanced expression of key innate immune antagonists Orf6, N and Orf9b in VOCs Alpha to Delta suggesting a not unusual evolutionary trajectory to combatting human innate immunity to beef up transmission1,2.Fig. 4: Innate immune phenotype of dominant Omicron subvariants. a, International SARS-CoV-2 variant collection counts over the years (scaled in keeping with variant), extracted from CoV-Spectrum the use of genomic knowledge from GISAID. b–d, Calu-3 cells have been contaminated with 2,000 Nsp12 copies in keeping with mobile. Replication of Omicron subvariants when compared with BA.2 (blue) and BA.5 (pink) measured by means of Nsp12 copies in keeping with microgram RNA is proven for BA.2.75 (yellow; Ο) (b), XBB subvariants (XBB.1: mild pink, Ο; XBB.1 (B): pink, Δ; XBB.1.5: darkish pink, □) (c) and BQ.1.1 (BQ.1.1: mild inexperienced, Ο; BQ.1.1 (B): darkish inexperienced, Δ) (d) isolates. e, HAEs have been contaminated with 1,500 Nsp12 copies in keeping with mobile and intracellular Nsp12 copies measured at 72 h.p.i. 3 organic replicates proven. f, IFNB and CXCL10 expression in Calu-3 cells contaminated with 2,000 Nsp12 copies in keeping with mobile of the indicated Omicron subvariants at 24 h.p.i. g, Viral replication of indicated variants in Calu-3 cells within the presence or absence of five μM ruxolitinib at 48 h.p.i. Numbers point out fold alternate in replication within the presence of five μM ruxolitinib. h,i, Western blot of Orf6, N, spike and β-actin at 48 h.p.i. in cells from b–d within the absence (h) or presence (i) of five μM ruxolitinib. j, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin in Calu-3 cells at 48 h.p.i. ok,l, Quantification of 2 impartial western blots of IRF3-pS396 (ok) and STAT1-pS727 (l) over β-actin at 24 h.p.i. For b–d, variant replication was once when compared with BA.2 at each and every timepoint the use of a two-way research of variance (ANOVA) and Bonferroni post-test. Colors point out comparator (BA.5, pink; BA.2.75, yellow; XBB.1, mild pink; XBB.1 (B), pink; XBB.1.5, darkish pink; BQ.1.1, mild inexperienced; BQ.1.1 (B), darkish inexperienced). For e–g, one-way ANOVA with Dunnett’s post-test was once used to match all variants with BA.2. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven. For f, ***P < 0.0001.Supply knowledge
a, International SARS-CoV-2 variant collection counts over the years (scaled in keeping with variant), extracted from CoV-Spectrum the use of genomic knowledge from GISAID. b–d, Calu-3 cells have been contaminated with 2,000 Nsp12 copies in keeping with mobile. Replication of Omicron subvariants when compared with BA.2 (blue) and BA.5 (pink) measured by means of Nsp12 copies in keeping with microgram RNA is proven for BA.2.75 (yellow; Ο) (b), XBB subvariants (XBB.1: mild pink, Ο; XBB.1 (B): pink, Δ; XBB.1.5: darkish pink, □) (c) and BQ.1.1 (BQ.1.1: mild inexperienced, Ο; BQ.1.1 (B): darkish inexperienced, Δ) (d) isolates. e, HAEs have been contaminated with 1,500 Nsp12 copies in keeping with mobile and intracellular Nsp12 copies measured at 72 h.p.i. 3 organic replicates proven. f, IFNB and CXCL10 expression in Calu-3 cells contaminated with 2,000 Nsp12 copies in keeping with mobile of the indicated Omicron subvariants at 24 h.p.i. g, Viral replication of indicated variants in Calu-3 cells within the presence or absence of five μM ruxolitinib at 48 h.p.i. Numbers point out fold alternate in replication within the presence of five μM ruxolitinib. h,i, Western blot of Orf6, N, spike and β-actin at 48 h.p.i. in cells from b–d within the absence (h) or presence (i) of five μM ruxolitinib. j, Western blot of STAT1-pY701, STAT1-pS727, general STAT1, IRF3-pS396, general IRF3 and β-actin in Calu-3 cells at 48 h.p.i. ok,l, Quantification of 2 impartial western blots of IRF3-pS396 (ok) and STAT1-pS727 (l) over β-actin at 24 h.p.i. For b–d, variant replication was once when compared with BA.2 at each and every timepoint the use of a two-way research of variance (ANOVA) and Bonferroni post-test. Colors point out comparator (BA.5, pink; BA.2.75, yellow; XBB.1, mild pink; XBB.1 (B), pink; XBB.1.5, darkish pink; BQ.1.1, mild inexperienced; BQ.1.1 (B), darkish inexperienced). For e–g, one-way ANOVA with Dunnett’s post-test was once used to match all variants with BA.2. Mirror measurements from one in all 3 impartial experiments. Fold alternate over mock is proven. Imply ± s.e.m. or particular person datapoints are proven. For f, ***P < 0.0001.Supply knowledge
Evolution of enhanced innate immune suppression by means of SARS-CoV-2 Omicron subvariants – Nature Microbiology















