The starting place and growth of cell-surface receptors within the plant lineagePlant cell-surface receptors which are identified to take part in immunity, advancement, and reproductive processes come with the LRR-, G-lectin-, Wall-associated kinase (WAK)-, Area of Unknown Serve as 26 (Duf26)-, L-lectin-, Lysin motif (LysM)-, and Malectin-containing RLKs and RLPs (Fig. 1a–h). There are further RLK households with other ectodomains, such because the proline-rich extensin-like receptor kinases (PERKs) and thaumatin-like protein kinases (TLPKs)9,13. Alternatively, their serve as in immunity isn’t well-characterized. Mobile-surface receptors with LRR-, G-lectin-, WAK-, and LysM-ectodomains had been reported to recognise PAMPs, whilst others understand self-molecules or unidentified ligands (Fig. 1h; Supplementary Fig. 1). Reputation of the varied array of ligands may be completed by way of variable buildings and mixtures of various ectodomains (Fig. 1a–g). To track the origins of various receptor categories throughout the plant lineage, we first recognized RLKs and RLPs in 350 genomes from Glaucophyta, crimson algae, inexperienced algae, Bryophytes, and Tracheophytes. We outline right here RLKs as any proteins with each 1–2 TMs and KDs, and RLPs as any protein with 1–2 TMs, however lack KDs. In overall, we recognized 177,645 RLKs, virtually as much as 70% of which possess both LRR-, G-lectin-, WAK-, Duf26-, L-lectin-, LysM- and Malectin-ectodomains (Fig. 1i). Subsequent, we looked for proteins with those ectodomains and TMs that lack KDs and located 41,144 RLPs (Fig. 1j). We additional tested which of the recognized RLKs and RLPs households usually are concerned about immunity. A prior record urged a favorable correlation between the gene circle of relatives sizes of cell-surface immune receptors and intracellular immune receptors (the NB-ARC circle of relatives) around the angiosperms4. We tested the correlation between the relative dimension (%; choice of recognized genes within the circle of relatives/numbers of searched genes × 100; see strategies) of the RLK households, the RLPs households, and the NB-ARC circle of relatives in each and every genome. Particularly, maximum RLK households (excluding for the LysM-RLKs) showcase certain correlations with the NB-ARC circle of relatives, whilst maximum RLP households (excluding for the LRR-RLPs) don’t showcase certain correlation with the NB-ARC circle of relatives (Primary Fig. 1k). Moreover, we checked the expression stage of those receptor households in Arabidopsis thaliana all over immunity. Particularly, the RLKs, excluding for LRR- and Malectin-RLKs, usually showcase upper expression ranges in comparison to the RLPs all over immunity (Primary Fig. 1k; Supplementary Fig. 2). Those information jointly counsel that the RLKs are much more likely to be concerned about immunity than the RLPs.Fig. 1: The distribution of cell-surface receptors in crops.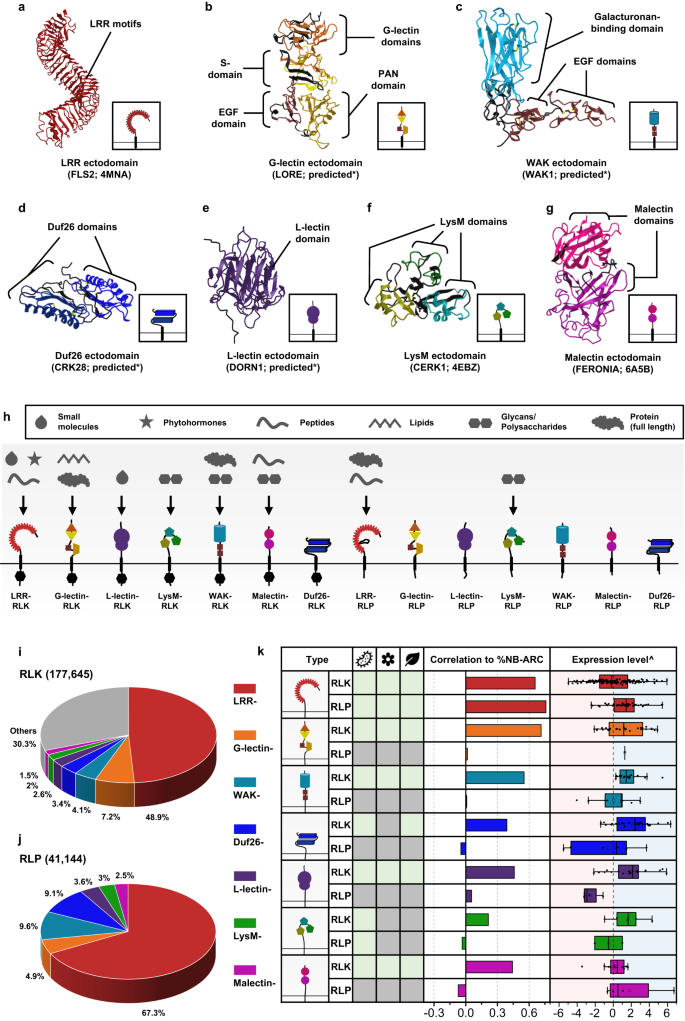 a–g Ectodomain construction of an LRR receptor. a a G-lectin receptor, b an L-lectin receptor, c a LysM receptor, d a Malectin receptor, e a WAK receptor, f and a Duf26 receptor g. Constructions of FLS2, CERK1, and FERONIA had been published55,95,96. Constructions of LORE, DORN1, WAK1 and CRK28 had been predicted by way of Alphafold2*97. Ectodomains are visualized in iCn3D98. h Schematic presentations the area structure of various categories of receptor-like kinase (RLKs) and receptor-like proteins (RLP) in crops. Arrows constitute the ligands that those receptor categories had been reported to understand or acknowledge. The higher field defines the ligands identified by way of other receptors. The decrease field defines the domain names throughout the receptor categories. Word that those receptors might be able to recognise different unidentified ligands. For more info, see Supplementary Fig. 1. i Ectodomain distribution of RLKs in crops. Every fraction represents the proportion (%) of ectodomain out of all of the RLKs from 350 species (177,645). j Ectodomain distribution of RLP in crops. Every fraction represents the proportion (%) of ectodomains out of all of the RLPs with the ones seven ectodomains (41,144). ok Desk of RLKs and RLPs with LRR (crimson), G-lectin (orange), WAK (turquoise), Duf26 (blue), L-lectin (crimson), LysM (inexperienced), and Malectin (magenta) ectodomains. Characterized receptors concerned about microbial interplay (micro organism icon), replica (flower icon), and advancement (leaf icon) are indicated with gentle inexperienced packing containers. Gray packing containers point out that the receptor elegance has no longer been reported to be concerned about that organic procedure. For main points, check with Supplementary Fig. 1. Correlations between other categories of cell-surface receptors and NB-ARC in 300 angiosperms are indicated with bars. Robust certain correlations are indicated by way of extension to the sunshine inexperienced house (Pearson’s r > 0.6) and medium certain correlations are throughout the yellow house (Pearson’s r between 0.3 and zero.6). Expression stage^ refers back to the expression of each and every elegance of cell-surface receptors all over NLR-triggered immunity (NTI) in Arabidopsis thaliana. Mild blue house represents larger expression and light-weight red house represents lowered expression all over NTI. X-axis values constitute log2 (fold exchange all over ETI relative to untreated samples). Boxplot components: middle line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. Collection of cell-surface receptors (n) analysed within the RNA-seq information: LRR-RLK, n = 159; LRR-RLP, n = 42; G-lectin-RLK, n = 29; G-lectin-RLP, n = 1; WAK-RLK, n = 18; WAK-RLP, n = 10; Duf26-RLK, n = 33; Duf26-RLP, n = 7; L-lectin-RLK, n = 21; L-lectin-RLP, n = 4; LysM-RLK, n = 5; LysM-RLP, n = 2; Malectin-RLK, n = 13; Malectin-RLP, n = 4. RNA-seq information analysed right here had been reported prior to now, the place NTI used to be activated by way of estradiol-induced expression of AvrRps4 in A. thaliana for 4 h94. For the expression of each and every elegance of cell-surface receptors all over PTI in A. thaliana, check with Supplementary Fig. 2.Subsequent, we tested the presence or absence of ectodomains (LRR-, G-lectin-, WAK-, Duf26-, L-lectin-, LysM- and Malectin-ectodomains missing TM or KD; ectodomain-only proteins), RLPs (TM-bound ectodomains) and RLKs (ectodomains encompassing each TM and KD) within the plant lineage (Fig. 2; Supplementary Fig. 3; Supplementary Information 1a–c). Ectodomains showcase an historical heritage, with LRR-, WAK-, LysM-, Malectin-, and L-lectin-domains courting again to the generation of Glaucophyta. In a similar fashion, fairly historical opposite numbers similar to LRR-RLPs, WAK-RLPs, LysM-RLPs, Malectin-RLPs, and L-lectin-RLPs are present in each Glaucophyta and Rhodophyta. By contrast, RLKs emerged extra just lately. Inexperienced algae harbour WAK-RLKs, Malectin-RLKs, and G-lectin-RLKs, and LysM-RLKs, L-lectin-RLKs, and Duf-26-RLKs are unique to Embryophytes (Fig. 2). Except for for LRR-RLPs, all six households of RLP are basal to the RLK households. This means the intriguing chance that some RLKs can have developed immediately from RLPs during the integration of kinase domain names. To check this speculation, we aligned the G-lectin-, WAK-, Duf26-, L-lectin-, LysM- and Malectin-ectodomains from the ectodomain-only proteins, RLPs, and RLKs inside of both a subset of 25 species or all 350 species (Supplementary Fig. 4). In some circumstances, we seen proportions of RLKs (similar to WAK-RLKs) that most probably have immediately developed from the RLPs (Supplementary Fig. 4a). In different circumstances (such because the LysM-RLKs; Supplementary Fig. 4c), it isn’t transparent whether or not the RLKs developed from an ectodomain-only protein or an RLP. Thus, we concluded that RLKs have emerged from both the RLPs or immediately from ectodomain-only proteins. Throughout the international ectodomain collection similarity timber, we additionally seen RLKs (the LysM-RLKs and Malectin-RLKs; Supplementary Fig. 4d, f) that experience emerged from two unbiased occasions. Whilst different RLKs (WAK-, G-lectin-, L-lectin- and Duf26-RLKs; Supplementary Fig. 4b, h, j, l) more likely to have emerged from RLPs or ectodomain-only proteins thru a sequence of domain-swapping occasions. Such occasions most probably befell at more than a few levels of plant evolution, given the presence of RLKs in various plant lineages14.Fig. 2: The starting place and growth of cell-surface receptors in crops.
a–g Ectodomain construction of an LRR receptor. a a G-lectin receptor, b an L-lectin receptor, c a LysM receptor, d a Malectin receptor, e a WAK receptor, f and a Duf26 receptor g. Constructions of FLS2, CERK1, and FERONIA had been published55,95,96. Constructions of LORE, DORN1, WAK1 and CRK28 had been predicted by way of Alphafold2*97. Ectodomains are visualized in iCn3D98. h Schematic presentations the area structure of various categories of receptor-like kinase (RLKs) and receptor-like proteins (RLP) in crops. Arrows constitute the ligands that those receptor categories had been reported to understand or acknowledge. The higher field defines the ligands identified by way of other receptors. The decrease field defines the domain names throughout the receptor categories. Word that those receptors might be able to recognise different unidentified ligands. For more info, see Supplementary Fig. 1. i Ectodomain distribution of RLKs in crops. Every fraction represents the proportion (%) of ectodomain out of all of the RLKs from 350 species (177,645). j Ectodomain distribution of RLP in crops. Every fraction represents the proportion (%) of ectodomains out of all of the RLPs with the ones seven ectodomains (41,144). ok Desk of RLKs and RLPs with LRR (crimson), G-lectin (orange), WAK (turquoise), Duf26 (blue), L-lectin (crimson), LysM (inexperienced), and Malectin (magenta) ectodomains. Characterized receptors concerned about microbial interplay (micro organism icon), replica (flower icon), and advancement (leaf icon) are indicated with gentle inexperienced packing containers. Gray packing containers point out that the receptor elegance has no longer been reported to be concerned about that organic procedure. For main points, check with Supplementary Fig. 1. Correlations between other categories of cell-surface receptors and NB-ARC in 300 angiosperms are indicated with bars. Robust certain correlations are indicated by way of extension to the sunshine inexperienced house (Pearson’s r > 0.6) and medium certain correlations are throughout the yellow house (Pearson’s r between 0.3 and zero.6). Expression stage^ refers back to the expression of each and every elegance of cell-surface receptors all over NLR-triggered immunity (NTI) in Arabidopsis thaliana. Mild blue house represents larger expression and light-weight red house represents lowered expression all over NTI. X-axis values constitute log2 (fold exchange all over ETI relative to untreated samples). Boxplot components: middle line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. Collection of cell-surface receptors (n) analysed within the RNA-seq information: LRR-RLK, n = 159; LRR-RLP, n = 42; G-lectin-RLK, n = 29; G-lectin-RLP, n = 1; WAK-RLK, n = 18; WAK-RLP, n = 10; Duf26-RLK, n = 33; Duf26-RLP, n = 7; L-lectin-RLK, n = 21; L-lectin-RLP, n = 4; LysM-RLK, n = 5; LysM-RLP, n = 2; Malectin-RLK, n = 13; Malectin-RLP, n = 4. RNA-seq information analysed right here had been reported prior to now, the place NTI used to be activated by way of estradiol-induced expression of AvrRps4 in A. thaliana for 4 h94. For the expression of each and every elegance of cell-surface receptors all over PTI in A. thaliana, check with Supplementary Fig. 2.Subsequent, we tested the presence or absence of ectodomains (LRR-, G-lectin-, WAK-, Duf26-, L-lectin-, LysM- and Malectin-ectodomains missing TM or KD; ectodomain-only proteins), RLPs (TM-bound ectodomains) and RLKs (ectodomains encompassing each TM and KD) within the plant lineage (Fig. 2; Supplementary Fig. 3; Supplementary Information 1a–c). Ectodomains showcase an historical heritage, with LRR-, WAK-, LysM-, Malectin-, and L-lectin-domains courting again to the generation of Glaucophyta. In a similar fashion, fairly historical opposite numbers similar to LRR-RLPs, WAK-RLPs, LysM-RLPs, Malectin-RLPs, and L-lectin-RLPs are present in each Glaucophyta and Rhodophyta. By contrast, RLKs emerged extra just lately. Inexperienced algae harbour WAK-RLKs, Malectin-RLKs, and G-lectin-RLKs, and LysM-RLKs, L-lectin-RLKs, and Duf-26-RLKs are unique to Embryophytes (Fig. 2). Except for for LRR-RLPs, all six households of RLP are basal to the RLK households. This means the intriguing chance that some RLKs can have developed immediately from RLPs during the integration of kinase domain names. To check this speculation, we aligned the G-lectin-, WAK-, Duf26-, L-lectin-, LysM- and Malectin-ectodomains from the ectodomain-only proteins, RLPs, and RLKs inside of both a subset of 25 species or all 350 species (Supplementary Fig. 4). In some circumstances, we seen proportions of RLKs (similar to WAK-RLKs) that most probably have immediately developed from the RLPs (Supplementary Fig. 4a). In different circumstances (such because the LysM-RLKs; Supplementary Fig. 4c), it isn’t transparent whether or not the RLKs developed from an ectodomain-only protein or an RLP. Thus, we concluded that RLKs have emerged from both the RLPs or immediately from ectodomain-only proteins. Throughout the international ectodomain collection similarity timber, we additionally seen RLKs (the LysM-RLKs and Malectin-RLKs; Supplementary Fig. 4d, f) that experience emerged from two unbiased occasions. Whilst different RLKs (WAK-, G-lectin-, L-lectin- and Duf26-RLKs; Supplementary Fig. 4b, h, j, l) more likely to have emerged from RLPs or ectodomain-only proteins thru a sequence of domain-swapping occasions. Such occasions most probably befell at more than a few levels of plant evolution, given the presence of RLKs in various plant lineages14.Fig. 2: The starting place and growth of cell-surface receptors in crops. The highest panel represents a chain similarity tree of a couple of algal and plant lineages. Circles (○) and stars (☆) point out the starting place and growth of receptor households. The timescale (in hundreds of thousands of years; MYA) of the collection similarity tree used to be estimated by way of TIMETREE599. The ground panel represents the presence or absence of various receptor categories in algal and plant lineages. ‘-’ represents ectodomains and not using a transmembrane or kinase area, ‘RLP’ represents ectodomains with a transmembrane area however no kinase area, ‘RLK’ represents ectodomains with each transmembrane and kinase domain names. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. The choice of species to be had from each and every algal and plant lineage is indicated by way of the numbers inside of respective packing containers. A gray field signifies the absence of receptors and a inexperienced field signifies their presence in each and every lineage. The starting place of a receptor is indicated with a circle (○). The origins of ‘-’, ‘RLP’, and ‘RLK’ are hooked up by way of black strains. Growth charges of receptor categories are indicated by way of boxplots. The chances (%) of cell-surface receptors from each and every genome had been calculated as (choice of recognized genes/choice of searched genes × 100). Subsequent, the odds from each and every species inside of a lineage (e.g., Rhodophtya or inexperienced algae) had been grouped and the median proportion used to be calculated. Median price used to be used as an alternative of imply to steer clear of outliers throughout the lineages. The growth charge inside of a species is calculated by way of ((% mobile floor receptors in that species)-(median))/(median). The cyan boxplot represents the growth charge from Glaucophyta and Rhodophyta to inexperienced algae (LRR, n = 20; LRR-RLP, n = 13; LRR-RLK, n = 9; WAK, n = 16; WAK-RLP, n = 19; WAK-RLK, n = 0; LysM, n = 20; LysM-RLP, n = 16; LysM-RLK, n = 0; Malectin, n = 9; Malectin-RLP, n = 6; Malectin-RLK, n = 0; G-lectin, n = 0; G-lectin-RLP, n = 0; G-lectin-RLK, n = 0; L-lectin, n = 8; L-lectin-RLP, n = 2; L-lectin-RLK, n = 0; Duf26, n = 0; Duf26-RLP, n = 0; Duf26-RLK, n = 0). The yellow boxplot represents the growth charge from inexperienced algae to Embryophytes (LRR, n = 324; LRR-RLP, n = 324; LRR-RLK, n = 324; WAK, n = 316; WAK-RLP, n = 324; WAK-RLK, n = 323; LysM, n = 323; LysM-RLP, n = 314; LysM-RLK, n = 0; Malectin, n = 321; Malectin-RLP, n = 294; Malectin-RLK, n = 319; G-lectin, n = 319; G-lectin-RLP, n = 315; G-lectin-RLK, n = 322; L-lectin, n = 277; L-lectin-RLP, n = 314; L-lectin-RLK, n = 0; Duf26, n = 0; Duf26-RLP, n = 0; Duf26-RLK, n = 0) and the orange boxplot represents the variations between early land crops to Tracheophytes (LRR, n = 314; LRR-RLP, n = 314; LRR-RLK, n = 314; WAK, n = 311; WAK-RLP, n = 314; WAK-RLK, n = 314; LysM, n = 313; LysM-RLP, n = 304; LysM-RLK, n = 312; Malectin, n = 311; Malectin-RLP, n = 292; Malectin-RLK, n = 311; G-lectin, n = 309; G-lectin-RLP, n = 307; G-lectin-RLK, n = 313; L-lectin, n = 267; L-lectin-RLP, n = 306; L-lectin-RLK, n = 312; Duf26, n = 312; Duf26-RLP, n = 313; Duf26-RLK, n = 0). Mild blue house represents growth and light-weight red house represents contraction of the gene circle of relatives. X-axis values constitute growth charge (×). Values better than 0 point out growth; values equivalent to 0 point out no growth, and values under 0 point out contraction. Boxplot components: centre line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. For main points, check with the strategies.We additionally tested the growth patterns of various receptor categories throughout more than a few plant lineages. Our research concerned calculating the median proportion (%) of cell-surface receptors households in (i) Glaucophyta and Rhodophyta, (ii) inexperienced algae, and (iii) Bryophytes to resolve the proportion building up (% building up; see strategies and Supplementary Word 2) from Glaucophyta and Rhodophyta to inexperienced algae; from inexperienced algae to Embryophytes; and between Bryophytes and Tracheophytes. We seen really extensive expansions in particular receptor households throughout those lineages. Inexperienced algae exhibited a vital growth of LRR-RLKs, whilst Embryophytes displayed expansions in LRR-RLPs, LRR-RLKs, WAK-RLKs, and G-lectin-RLKs. Tracheophytes had additional expansions in LRR-RLPs, WAK-RLKs, Malectin-RLKs, G-lectin-RLKs, and Duf26-RLPs (Fig. 2; Supplementary Fig. 3). Total, RLKs show higher growth in comparison to RLPs, with notable expansions seen in LRR-RLK, WAK-RLK, and G-lectin-RLK. As well as, the LRR-RLP circle of relatives has additionally considerably expanded during the plant lineage. Those findings align with the really extensive dimension of those receptor households and their involvement in recognising pathogens (Fig. 1i; Supplementary Fig. 1b). LRR-RLKs are labeled into 20 subgroups in keeping with their kinase domain names, with subgroup XII in particular implicated in PAMP recognition15. Specifically, the LRR-RLK-XII subgroup reveals a significantly upper growth charge in comparison to different subgroups (Supplementary Fig. 5), reinforcing the concept that mobile floor immune receptors underwent in depth expansions because the plant lineage assorted and developed to conform to a variety of environments.The starting place and growth of PTI-signalling elements within the plant lineageUpon ligand or elicitor belief, PRRs go through dimerisation or shape heteromeric complexes with different LRR-RLK co-receptors, similar to BAK1 (a member of the Somatic embryogenesis receptor-like kinases (SERK) circle of relatives) and SOBIR1. This spatial association brings the cytoplasmic kinase domain names (from co-receptors and/or receptors) in shut proximity, beginning a cascade of auto- and trans-phosphorylation events16. The activated receptor complicated due to this fact phosphorylates participants of the cytoplasmic receptor-like kinases subgroup VII (RLCK-VII)17, which, in flip, phosphorylate more than a few cytoplasmic kinases, such because the mitogen-activated protein kinase kinase kinases (MAPKKKs), calcium‐dependent protein kinases (CDPKs) and plasma membrane-associated proteins, similar to cyclic nucleotide-gated channels (CNGCs), hyperosmolality-gated calcium-permeable channels (OSCAs), and NADPH oxidases (RBOHs)16. The phosphorylation of those proteins jointly triggers transcriptional reprogramming and physiological adjustments, similar to cytoplasmic calcium inflow and the buildup of reactive oxygen species (ROS)18. Those physiological responses successfully impede pathogen proliferation all over an infection (Fig. 2a; Supplementary Figs. 6 and seven).We recognized cell-surface co-receptors and signalling elements from the 350 genomes and made up our minds their absence or presence around the plant lineage (Supplementary Fig. 7). SERKs, appearing as cell-surface co-receptors for a couple of LRR-RLKs and LRR-RLPs are found in Zygnematophyceae and Embryophytes19 (Fig. 3b; Supplementary Fig. 8a, b), suggesting their emergence all over or previous to the illusion of land crops. Immune-related LRR-RLPs lack intracellular kinase domain names, thus require any other LRR-RLK co-receptor, SOBIR1, to turn on downstream signalling20. Very similar to BAK1, SOBIR1 may be found in Embryophytes (Fig. 3b). Thus, co-receptors for cell-surface receptors most probably developed all over or prior to the emergence of land crops. Then again, cytoplasmic kinases (RLCKs, CDPKs, MAPKKKs, MAPKKs, and MAPKs) are historical, as are the PM-localised downstream signalling elements (CNGCs, OCSAs, and RBOHs), discovered throughout all plant lineages (Fig. 3b; Supplementary Fig. 8). Despite the fact that the precise serve as of those proteins in algal species stays unclear, their immune-related orthologs are found in inexperienced algae (Fig. 3b; Supplementary Fig. 8. This means that they underwent specialisation throughout the immune activation pathway previous to the emergence of land crops. The EP proteins (EDS1, PAD4, and SAG101) and RPW8-NLRs (NRG1 and ADR1) which are very important for each TIR-NLR and LRR-RLP mediated-immunity21,22, are solely found in gymnosperms and angiosperms (seed crops)23. Taking into account the traditional nature of the LRR-RLPs, it’s believable that EP-protein and helper-NLRs had been built-in into the LRR-RLP-signalling pathway, forming a powerful immune community in seed crops.Fig. 3: The starting place and evolution of cell-surface receptor signalling element in crops.
The highest panel represents a chain similarity tree of a couple of algal and plant lineages. Circles (○) and stars (☆) point out the starting place and growth of receptor households. The timescale (in hundreds of thousands of years; MYA) of the collection similarity tree used to be estimated by way of TIMETREE599. The ground panel represents the presence or absence of various receptor categories in algal and plant lineages. ‘-’ represents ectodomains and not using a transmembrane or kinase area, ‘RLP’ represents ectodomains with a transmembrane area however no kinase area, ‘RLK’ represents ectodomains with each transmembrane and kinase domain names. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. The choice of species to be had from each and every algal and plant lineage is indicated by way of the numbers inside of respective packing containers. A gray field signifies the absence of receptors and a inexperienced field signifies their presence in each and every lineage. The starting place of a receptor is indicated with a circle (○). The origins of ‘-’, ‘RLP’, and ‘RLK’ are hooked up by way of black strains. Growth charges of receptor categories are indicated by way of boxplots. The chances (%) of cell-surface receptors from each and every genome had been calculated as (choice of recognized genes/choice of searched genes × 100). Subsequent, the odds from each and every species inside of a lineage (e.g., Rhodophtya or inexperienced algae) had been grouped and the median proportion used to be calculated. Median price used to be used as an alternative of imply to steer clear of outliers throughout the lineages. The growth charge inside of a species is calculated by way of ((% mobile floor receptors in that species)-(median))/(median). The cyan boxplot represents the growth charge from Glaucophyta and Rhodophyta to inexperienced algae (LRR, n = 20; LRR-RLP, n = 13; LRR-RLK, n = 9; WAK, n = 16; WAK-RLP, n = 19; WAK-RLK, n = 0; LysM, n = 20; LysM-RLP, n = 16; LysM-RLK, n = 0; Malectin, n = 9; Malectin-RLP, n = 6; Malectin-RLK, n = 0; G-lectin, n = 0; G-lectin-RLP, n = 0; G-lectin-RLK, n = 0; L-lectin, n = 8; L-lectin-RLP, n = 2; L-lectin-RLK, n = 0; Duf26, n = 0; Duf26-RLP, n = 0; Duf26-RLK, n = 0). The yellow boxplot represents the growth charge from inexperienced algae to Embryophytes (LRR, n = 324; LRR-RLP, n = 324; LRR-RLK, n = 324; WAK, n = 316; WAK-RLP, n = 324; WAK-RLK, n = 323; LysM, n = 323; LysM-RLP, n = 314; LysM-RLK, n = 0; Malectin, n = 321; Malectin-RLP, n = 294; Malectin-RLK, n = 319; G-lectin, n = 319; G-lectin-RLP, n = 315; G-lectin-RLK, n = 322; L-lectin, n = 277; L-lectin-RLP, n = 314; L-lectin-RLK, n = 0; Duf26, n = 0; Duf26-RLP, n = 0; Duf26-RLK, n = 0) and the orange boxplot represents the variations between early land crops to Tracheophytes (LRR, n = 314; LRR-RLP, n = 314; LRR-RLK, n = 314; WAK, n = 311; WAK-RLP, n = 314; WAK-RLK, n = 314; LysM, n = 313; LysM-RLP, n = 304; LysM-RLK, n = 312; Malectin, n = 311; Malectin-RLP, n = 292; Malectin-RLK, n = 311; G-lectin, n = 309; G-lectin-RLP, n = 307; G-lectin-RLK, n = 313; L-lectin, n = 267; L-lectin-RLP, n = 306; L-lectin-RLK, n = 312; Duf26, n = 312; Duf26-RLP, n = 313; Duf26-RLK, n = 0). Mild blue house represents growth and light-weight red house represents contraction of the gene circle of relatives. X-axis values constitute growth charge (×). Values better than 0 point out growth; values equivalent to 0 point out no growth, and values under 0 point out contraction. Boxplot components: centre line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. For main points, check with the strategies.We additionally tested the growth patterns of various receptor categories throughout more than a few plant lineages. Our research concerned calculating the median proportion (%) of cell-surface receptors households in (i) Glaucophyta and Rhodophyta, (ii) inexperienced algae, and (iii) Bryophytes to resolve the proportion building up (% building up; see strategies and Supplementary Word 2) from Glaucophyta and Rhodophyta to inexperienced algae; from inexperienced algae to Embryophytes; and between Bryophytes and Tracheophytes. We seen really extensive expansions in particular receptor households throughout those lineages. Inexperienced algae exhibited a vital growth of LRR-RLKs, whilst Embryophytes displayed expansions in LRR-RLPs, LRR-RLKs, WAK-RLKs, and G-lectin-RLKs. Tracheophytes had additional expansions in LRR-RLPs, WAK-RLKs, Malectin-RLKs, G-lectin-RLKs, and Duf26-RLPs (Fig. 2; Supplementary Fig. 3). Total, RLKs show higher growth in comparison to RLPs, with notable expansions seen in LRR-RLK, WAK-RLK, and G-lectin-RLK. As well as, the LRR-RLP circle of relatives has additionally considerably expanded during the plant lineage. Those findings align with the really extensive dimension of those receptor households and their involvement in recognising pathogens (Fig. 1i; Supplementary Fig. 1b). LRR-RLKs are labeled into 20 subgroups in keeping with their kinase domain names, with subgroup XII in particular implicated in PAMP recognition15. Specifically, the LRR-RLK-XII subgroup reveals a significantly upper growth charge in comparison to different subgroups (Supplementary Fig. 5), reinforcing the concept that mobile floor immune receptors underwent in depth expansions because the plant lineage assorted and developed to conform to a variety of environments.The starting place and growth of PTI-signalling elements within the plant lineageUpon ligand or elicitor belief, PRRs go through dimerisation or shape heteromeric complexes with different LRR-RLK co-receptors, similar to BAK1 (a member of the Somatic embryogenesis receptor-like kinases (SERK) circle of relatives) and SOBIR1. This spatial association brings the cytoplasmic kinase domain names (from co-receptors and/or receptors) in shut proximity, beginning a cascade of auto- and trans-phosphorylation events16. The activated receptor complicated due to this fact phosphorylates participants of the cytoplasmic receptor-like kinases subgroup VII (RLCK-VII)17, which, in flip, phosphorylate more than a few cytoplasmic kinases, such because the mitogen-activated protein kinase kinase kinases (MAPKKKs), calcium‐dependent protein kinases (CDPKs) and plasma membrane-associated proteins, similar to cyclic nucleotide-gated channels (CNGCs), hyperosmolality-gated calcium-permeable channels (OSCAs), and NADPH oxidases (RBOHs)16. The phosphorylation of those proteins jointly triggers transcriptional reprogramming and physiological adjustments, similar to cytoplasmic calcium inflow and the buildup of reactive oxygen species (ROS)18. Those physiological responses successfully impede pathogen proliferation all over an infection (Fig. 2a; Supplementary Figs. 6 and seven).We recognized cell-surface co-receptors and signalling elements from the 350 genomes and made up our minds their absence or presence around the plant lineage (Supplementary Fig. 7). SERKs, appearing as cell-surface co-receptors for a couple of LRR-RLKs and LRR-RLPs are found in Zygnematophyceae and Embryophytes19 (Fig. 3b; Supplementary Fig. 8a, b), suggesting their emergence all over or previous to the illusion of land crops. Immune-related LRR-RLPs lack intracellular kinase domain names, thus require any other LRR-RLK co-receptor, SOBIR1, to turn on downstream signalling20. Very similar to BAK1, SOBIR1 may be found in Embryophytes (Fig. 3b). Thus, co-receptors for cell-surface receptors most probably developed all over or prior to the emergence of land crops. Then again, cytoplasmic kinases (RLCKs, CDPKs, MAPKKKs, MAPKKs, and MAPKs) are historical, as are the PM-localised downstream signalling elements (CNGCs, OCSAs, and RBOHs), discovered throughout all plant lineages (Fig. 3b; Supplementary Fig. 8). Despite the fact that the precise serve as of those proteins in algal species stays unclear, their immune-related orthologs are found in inexperienced algae (Fig. 3b; Supplementary Fig. 8. This means that they underwent specialisation throughout the immune activation pathway previous to the emergence of land crops. The EP proteins (EDS1, PAD4, and SAG101) and RPW8-NLRs (NRG1 and ADR1) which are very important for each TIR-NLR and LRR-RLP mediated-immunity21,22, are solely found in gymnosperms and angiosperms (seed crops)23. Taking into account the traditional nature of the LRR-RLPs, it’s believable that EP-protein and helper-NLRs had been built-in into the LRR-RLP-signalling pathway, forming a powerful immune community in seed crops.Fig. 3: The starting place and evolution of cell-surface receptor signalling element in crops. a Schematic determine represents the simplified PTI signalling pathway in crops. Colored hexagons on RLKs point out activated kinases. For main points, check with Supplementary Fig. 6. b The highest panel is a chain similarity tree of a couple of algal and plant lineages. Circles (○) and stars (☆) point out the origins and growth of receptor households, respectively. The timescale (in million years; MYA) of the collection similarity tree used to be estimated by way of TIMETREE599. The ground panel presentations the presence or absence of receptor categories in numerous algal and plant lineages. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. The choice of to be had species from each and every algal and plant lineage is indicated throughout the respective packing containers. A gray field signifies the absence, whilst a inexperienced field signifies the presence of a given protein circle of relatives in each and every lineage. Darkish inexperienced signifies the presence of orthologs of immunity-related (PTI) signalling elements inside of that protein circle of relatives (see additionally Supplementary Fig. 8). The starting place of a protein circle of relatives is indicated with a circle (○), adopted by way of any other circle indicating the starting place of the orthologs of PTI-signalling element. Growth charges of PTI-signalling element households are indicated by way of boxplots. The chances (%) of signalling elements from each and every genome had been calculated as (choice of recognized genes/choice of searched genes × 100). Subsequent, the odds from each and every species inside of a lineage (e.g, Rhodophtya or inexperienced algae) had been grouped and the median proportion used to be calculated. Median price used to be used as an alternative of imply to steer clear of outliers throughout the lineages. The growth charge inside of a species is calculated by way of ((%signalling elements in that species)-(median))/(median). The cyan boxplot represents the growth charge from Glaucophyta and Rhodophyta to inexperienced algae (SERKs, n = 0; SOBIR1, n = 0; RLKCs (VII), n = 0; CDPKs, n = 20; MAPKKKs, n = 20; MAPKKs, n = 20; MAPKs, n = 20; CNGCs, n = 19; OSCAs, n = 19; RBOHs, n = 10; EP proteins, n = 0; RPW8-NLRs, n = 0). The yellow boxplot represents the growth charge from inexperienced algae to Embryophytes (SERKs, n = 316; SOBIR1, n = 0; RLKCs (VII), n = 324; CDPKs, n = 324; MAPKKKs, n = 324; MAPKKs, n = 324; MAPKs, n = 324; CNGCs, n = 324; OSCAs, n = 322; RBOHs, n = 324; EP proteins, n = 0; RPW8-NLRs, n = 0) and the orange boxplot represents the variations between early land crops to Tracheophytes (SERKs, n = 307; SOBIR1, n = 309; RLKCs (VII), n = 314; CDPKs, n = 314; MAPKKKs, n = 314; MAPKKs, n = 314; MAPKs, n = 314; CNGCs, n = 314; OSCAs, n = 312; RBOHs, n = 314; EP proteins, n = 0; RPW8-NLRs, n = 0). Mild blue house represents growth and light-weight red house represents contraction of the gene circle of relatives. X-axis values constitute growth charge (×). Values better than 0 point out growth; values equivalent to 0 point out no growth, and values under 0 point out contraction. Boxplot components: centre line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. For main points, check with the strategies.Our investigation of the growth charge of signalling elements throughout the plant lineage indicated a diffusion of CDPKs in inexperienced algae and growth of RLCK-VIIs in Tracheophytes (Fig. 3b; Supplementary Fig. 8; Supplementary Word 2). Alternatively, different households of signalling elements showcase extra restricted expansions, in comparison to cell-surface receptors. This may be in step with the significantly better circle of relatives sizes of cell-surface receptors and NLRs compared to the signalling elements (Supplementary Fig. 9). Moreover, we tested the correlation between the odds of signalling elements and PRRs (LRR-RLK-XIIs + LRR-RLPs) throughout genomes. Except for for CNGCs, EP proteins, and RPW8-NLRs (0.6 > Pearson’s r > 0.3), maximum signalling element households don’t showcase co-expansion or co-contraction with PRRs (Supplementary Fig. 9). Thus, we concluded that crops are much more likely to conform new receptors relatively than downstream signalling elements for adaptation. The RLCK-VIIs are additional labeled into ten subgroups that are differentially required for RLKs and RLPs to turn on downstream responses17,24,25,26,27 (Supplementary Figs. 6 and seven). In a similar fashion, CDPKs fall into 4 subgroups (Supplementary Fig. 8). RLCK-VII and CDPK subgroup participants are differentially required by way of other PRRs to turn on downstream responses28,29,30,31. Pathogens incessantly goal RLCK-VIIs thru secreted effectors to suppress immunity32,33,34. Thus, redundancy amongst RLCK-VII subgroups serves as a protecting mechanism for the downstream signalling pathway towards effector focused on. As well as, crops have developed RLCK-VII pseudokinases, or ‘decoys’, to protect purposeful RLCKs thru NLRs34,35,36,37,38. In combination, it has change into obvious that the growth of RLCK-VII households can have been pushed by way of pathogenic power, thereby contributing to the improved robustness of the immune signalling community.Immunity- and development-related mobile floor receptors percentage a not unusual originWe sought to know how cell-surface receptors developed to be specialized in immunity. To reach this, we made up our minds to track the evolutionary starting place of LRR-RLPs in crops. Amongst RLPs, LRR-RLPs represent the biggest circle of relatives, comprising greater than twenty characterized participants that understand PAMPs or apoplastic effectors to turn on immunity (Supplementary Fig. 10). The ectodomain of LRR-RLPs encompasses further domain names referred to as N-loop outs (NLs) and island domain names (IDs) interspersed between the LRR motifs39,40. In most cases, NLs are positioned nearer to the N-termini of the ectodomain, while IDs are located nearer to C-termini39,40. NLs are found in maximum immunity-related LRR-RLPs whilst IDs are found in all immunity-related LRR-RLPs (Supplementary Figs. 10 and 11). NL positioning is fairly extra versatile, happening both prior to the primary LRR motif or between the primary few LRR motifs. Conversely, ID positioning is much less versatile, positioned most commonly prior to the final 4 LRR motifs throughout the ectodomain (Supplementary Figs. 10 and 11). This remark implies a purposeful necessity for the precise placement of IDs in LRR-RLPs.To research the purposeful necessity of IDs, we analysed ectodomains of LRR-RLPs and LRR-RLKs from 350 species (113,794) (Fig. 4a and Supplementary Fig. 12a, b). Using a couple of prediction methods, we recognized gaps between LRR motifs starting from 10–29 or 30–90 amino acids (AA) (Supplementary Fig. 12a). Since NLs normally span 6–30AA and IDs vary from round 40–75AA, we interested in small gaps (10–29AA) akin to NLs, and big gaps (30–90AA) indicative of IDs (Supplementary Fig. 10). Small or huge gaps are fairly rare in LRR-RLKs (10.6% and 5.43%, respectively) (Fig. 4a). By contrast, each small and big gaps are extra prevalent in LRR-RLPs (28.3% and 61.6%, respectively). Moreover, each LRR-RLKs and LRR-RLPs normally have just one hole, which can also be both small or huge (Supplementary Fig. 12c, d). Our research additionally confirmed that small hole positions throughout the ectodomains of each LRR-RLKs and LRR-RLPs don’t seem to be mounted, however is also disbursed randomly. Conversely, better gaps are predominantly located prior to the final 4 LRR motifs within the ectodomain (51.2% for LRR-RLKs and 86.9% for LRR-RLPs) (Fig. 4b, c and Supplementary Fig. 13). Thus, our findings counsel purposeful requirement for IDs to be located prior to the final 4 LRRs.Fig. 4: The starting place and evolution of LRRID + 4LRR in crops.
a Schematic determine represents the simplified PTI signalling pathway in crops. Colored hexagons on RLKs point out activated kinases. For main points, check with Supplementary Fig. 6. b The highest panel is a chain similarity tree of a couple of algal and plant lineages. Circles (○) and stars (☆) point out the origins and growth of receptor households, respectively. The timescale (in million years; MYA) of the collection similarity tree used to be estimated by way of TIMETREE599. The ground panel presentations the presence or absence of receptor categories in numerous algal and plant lineages. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. The choice of to be had species from each and every algal and plant lineage is indicated throughout the respective packing containers. A gray field signifies the absence, whilst a inexperienced field signifies the presence of a given protein circle of relatives in each and every lineage. Darkish inexperienced signifies the presence of orthologs of immunity-related (PTI) signalling elements inside of that protein circle of relatives (see additionally Supplementary Fig. 8). The starting place of a protein circle of relatives is indicated with a circle (○), adopted by way of any other circle indicating the starting place of the orthologs of PTI-signalling element. Growth charges of PTI-signalling element households are indicated by way of boxplots. The chances (%) of signalling elements from each and every genome had been calculated as (choice of recognized genes/choice of searched genes × 100). Subsequent, the odds from each and every species inside of a lineage (e.g, Rhodophtya or inexperienced algae) had been grouped and the median proportion used to be calculated. Median price used to be used as an alternative of imply to steer clear of outliers throughout the lineages. The growth charge inside of a species is calculated by way of ((%signalling elements in that species)-(median))/(median). The cyan boxplot represents the growth charge from Glaucophyta and Rhodophyta to inexperienced algae (SERKs, n = 0; SOBIR1, n = 0; RLKCs (VII), n = 0; CDPKs, n = 20; MAPKKKs, n = 20; MAPKKs, n = 20; MAPKs, n = 20; CNGCs, n = 19; OSCAs, n = 19; RBOHs, n = 10; EP proteins, n = 0; RPW8-NLRs, n = 0). The yellow boxplot represents the growth charge from inexperienced algae to Embryophytes (SERKs, n = 316; SOBIR1, n = 0; RLKCs (VII), n = 324; CDPKs, n = 324; MAPKKKs, n = 324; MAPKKs, n = 324; MAPKs, n = 324; CNGCs, n = 324; OSCAs, n = 322; RBOHs, n = 324; EP proteins, n = 0; RPW8-NLRs, n = 0) and the orange boxplot represents the variations between early land crops to Tracheophytes (SERKs, n = 307; SOBIR1, n = 309; RLKCs (VII), n = 314; CDPKs, n = 314; MAPKKKs, n = 314; MAPKKs, n = 314; MAPKs, n = 314; CNGCs, n = 314; OSCAs, n = 312; RBOHs, n = 314; EP proteins, n = 0; RPW8-NLRs, n = 0). Mild blue house represents growth and light-weight red house represents contraction of the gene circle of relatives. X-axis values constitute growth charge (×). Values better than 0 point out growth; values equivalent to 0 point out no growth, and values under 0 point out contraction. Boxplot components: centre line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles. For main points, check with the strategies.Our investigation of the growth charge of signalling elements throughout the plant lineage indicated a diffusion of CDPKs in inexperienced algae and growth of RLCK-VIIs in Tracheophytes (Fig. 3b; Supplementary Fig. 8; Supplementary Word 2). Alternatively, different households of signalling elements showcase extra restricted expansions, in comparison to cell-surface receptors. This may be in step with the significantly better circle of relatives sizes of cell-surface receptors and NLRs compared to the signalling elements (Supplementary Fig. 9). Moreover, we tested the correlation between the odds of signalling elements and PRRs (LRR-RLK-XIIs + LRR-RLPs) throughout genomes. Except for for CNGCs, EP proteins, and RPW8-NLRs (0.6 > Pearson’s r > 0.3), maximum signalling element households don’t showcase co-expansion or co-contraction with PRRs (Supplementary Fig. 9). Thus, we concluded that crops are much more likely to conform new receptors relatively than downstream signalling elements for adaptation. The RLCK-VIIs are additional labeled into ten subgroups that are differentially required for RLKs and RLPs to turn on downstream responses17,24,25,26,27 (Supplementary Figs. 6 and seven). In a similar fashion, CDPKs fall into 4 subgroups (Supplementary Fig. 8). RLCK-VII and CDPK subgroup participants are differentially required by way of other PRRs to turn on downstream responses28,29,30,31. Pathogens incessantly goal RLCK-VIIs thru secreted effectors to suppress immunity32,33,34. Thus, redundancy amongst RLCK-VII subgroups serves as a protecting mechanism for the downstream signalling pathway towards effector focused on. As well as, crops have developed RLCK-VII pseudokinases, or ‘decoys’, to protect purposeful RLCKs thru NLRs34,35,36,37,38. In combination, it has change into obvious that the growth of RLCK-VII households can have been pushed by way of pathogenic power, thereby contributing to the improved robustness of the immune signalling community.Immunity- and development-related mobile floor receptors percentage a not unusual originWe sought to know how cell-surface receptors developed to be specialized in immunity. To reach this, we made up our minds to track the evolutionary starting place of LRR-RLPs in crops. Amongst RLPs, LRR-RLPs represent the biggest circle of relatives, comprising greater than twenty characterized participants that understand PAMPs or apoplastic effectors to turn on immunity (Supplementary Fig. 10). The ectodomain of LRR-RLPs encompasses further domain names referred to as N-loop outs (NLs) and island domain names (IDs) interspersed between the LRR motifs39,40. In most cases, NLs are positioned nearer to the N-termini of the ectodomain, while IDs are located nearer to C-termini39,40. NLs are found in maximum immunity-related LRR-RLPs whilst IDs are found in all immunity-related LRR-RLPs (Supplementary Figs. 10 and 11). NL positioning is fairly extra versatile, happening both prior to the primary LRR motif or between the primary few LRR motifs. Conversely, ID positioning is much less versatile, positioned most commonly prior to the final 4 LRR motifs throughout the ectodomain (Supplementary Figs. 10 and 11). This remark implies a purposeful necessity for the precise placement of IDs in LRR-RLPs.To research the purposeful necessity of IDs, we analysed ectodomains of LRR-RLPs and LRR-RLKs from 350 species (113,794) (Fig. 4a and Supplementary Fig. 12a, b). Using a couple of prediction methods, we recognized gaps between LRR motifs starting from 10–29 or 30–90 amino acids (AA) (Supplementary Fig. 12a). Since NLs normally span 6–30AA and IDs vary from round 40–75AA, we interested in small gaps (10–29AA) akin to NLs, and big gaps (30–90AA) indicative of IDs (Supplementary Fig. 10). Small or huge gaps are fairly rare in LRR-RLKs (10.6% and 5.43%, respectively) (Fig. 4a). By contrast, each small and big gaps are extra prevalent in LRR-RLPs (28.3% and 61.6%, respectively). Moreover, each LRR-RLKs and LRR-RLPs normally have just one hole, which can also be both small or huge (Supplementary Fig. 12c, d). Our research additionally confirmed that small hole positions throughout the ectodomains of each LRR-RLKs and LRR-RLPs don’t seem to be mounted, however is also disbursed randomly. Conversely, better gaps are predominantly located prior to the final 4 LRR motifs within the ectodomain (51.2% for LRR-RLKs and 86.9% for LRR-RLPs) (Fig. 4b, c and Supplementary Fig. 13). Thus, our findings counsel purposeful requirement for IDs to be located prior to the final 4 LRRs.Fig. 4: The starting place and evolution of LRRID + 4LRR in crops.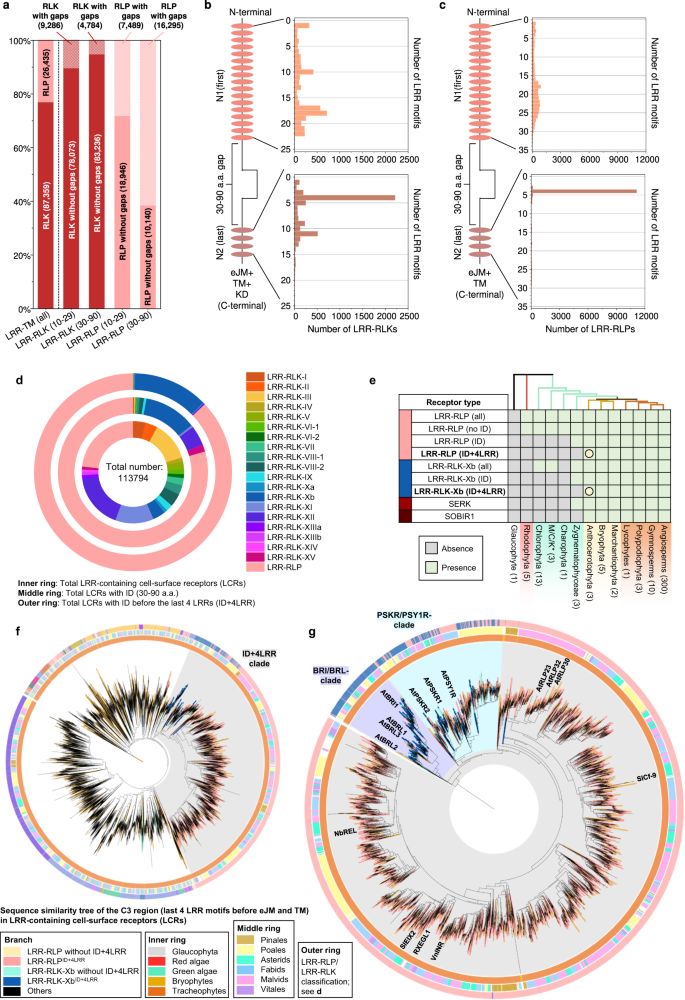 a Distribution of LRR-RLKs and LRR-RLPs in LRR-containing mobile floor receptors (LRR-TM) from 350 plant species, and the distribution of LRR-RLPs and LRR-RLKs without or with gaps of 10-29 amino acids (10-29) or 30-90 (30-90) amino acids. b, c Place of huge gaps (IDs; 30–90 AA) in b LRR-RLKs and c LRR-RLPs with a unmarried huge ID. N1 represents the choice of LRR motifs prior to the IDs and N2 represents the choice of LRR motifs after the ID. For positions of gaps in LRR-RLKs and LRR-RLPs with a couple of gaps, check with Supplementary Fig. 13. d The concentric ring pie chart gifts the proportion of LRR-containing cell-surface receptors (PRRs) from 350 species. The internal ring represents all LRR-containing cell-surface receptors (113,794); the center ring represents LRR-containing PRRs with ID (20,556); the outer ring represents LRR-containing PRRs with an ID previous the final 4 LRR (ID + 4LRR) on the C terminus (16,885). e The presence or absence of receptor categories in more than a few algal and plant lineages. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. A gray field signifies the absence, and a inexperienced field signifies the presence of a given receptor elegance in each and every lineage. The starting place of LRR-RLP and LRR-RLK-Xb with ID + 4LRR is indicated with a circle (○). f Series similarity tree of the C3 area (final 4 LRRs) from all LRR-containing cell-surface receptors of 350 species. Branches are colour-labelled as indicated. The internal ring and heart ring point out the lineage and subclass/order of the corresponding protein (species) from the department. Outer ring represents the LRR-RLP or LRR-RLK classification, which is indicated in d. The sunshine gray house signifies clustering of LRR-RLK-Xb and LRR-RLP with ID + 4LRR. The pruned collection similarity tree at the proper g corresponds to the sunshine gray house within the left tree, with clades labelled in darkish gray spaces accordingly. Characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade and the PSKR/PSY1R-clades also are labelled.Research of the distribution of LRR-RLK subgroups with IDs indicated that over 55% belong to the Xb subgroup (Fig. 4d). Moreover, 94.7% of LRR-RLKs with IDs located prior to the final 4 LRRs (ID + 4LRR) belong to the Xb subgroup (Fig. 4d), suggesting that each LRR-RLK-Xb and LRR-RLPs percentage the ID + 4LRR motif. A few of the characterized participants of LRR-RLK-Xb are essential elements of enlargement and advancement law, together with the BRASSINOSTEROID INSENSITIVE 1 (BRI1) members of the family, PHYTOSULFOKIN RECEPTOR 1 (PSKR1) and PSY1 RECEPTOR (PSY1R) members of the family, EXCESS MICROSPOROCYTES1 (EMS1) and NEMATODE-INDUCED LRR-RLK 1 (NILR1/GRACE)41,42,43,44,45. Curiously, we seen that each LRR-RLPs and LRR-RLK-Xbs with ID + 4LRR motifs are found in land crops (Embryophyte) however no longer in different lineages (Fig. 4e). Taking into account the similarity in structural motifs between LRR-RLPs and LRR-RLK-Xbs, it’s most probably that those two receptor categories percentage a not unusual starting place.To check this speculation, we carried out phylogenetic and structural analyses at the ectodomains of LRR-RLPs and LRR-RLKs. First, we aligned the final 4 LRR motifs (known as the C3 region46) from LRR-RLKs and LRR-RLPs (60,240). LRR-RLPs and LRR-RLK-Xbs with an ID + 4LRR cluster in combination within the C3 collection similarity tree, indicating a not unusual starting place (Fig. 4f). Inside this cluster (indicated because the ID + 4LRR clade), we seen distinct subclades, together with the BRI1/BRI1-LIKE (BRL) clade, the PSKR/PSY1R clade, and most commonly LRR-RLPs (Fig. 4g). The carefully equivalent C3 areas point out a conserved serve as. BRI1 recognises brassinosteroids (BRs) and interacts with the co-receptor BAK1 to urge BR responses47,48. In a similar fashion, PSKR1 perceives phytosulfokine (PSK) and interacts with the co-receptor SERK1/2. LRR-RLPs, then again, understand PAMPs or apoplastic effectors and interact each co-receptor BAK1 and SOBIR1 to start up immune responses49. Structural research have elucidated the interplay mechanism of BRI1, PSKR1, and the LRR-RLP RXEG1 with SERKs48,50,51,52,53,54 (Fig. 5a–c, Supplementary Fig. 14). Curiously, the C3 areas of BRI1, PSKR1, and RXEG1 play a a very powerful position in SERK interactions48,50,52,53, differentiating them from different LRR-RLKs. BRI1, PSKR1 and RXEG1 comprise particular amino acid residues throughout the terminal two LRR motifs (throughout the C3 area) to have interaction with BAK1/SERK1 (Fig. 5a–c), whilst the flagellin peptide (flg22) receptor FLS2 (an LRR-RLK-XII), is determined by the third–twelfth final LRR for BAK1 (SERK3) interactions55 (Supplementary Fig. 14a). The N-terminal area (or N-terminal cap) of BAK1 or SERK1 is essential for FLS2, BRI1, PSKR1, and RXEG1 interactions, whilst the first LRR internal floor of BAK1 or SERK1 is basically concerned about BRI1, PSKR1, and RXEG1 interactions, and the second and the 4th LRR internal floor of BAK1 is concerned about FLS2 interactions (Supplementary Fig. 14b). There also are putting similarities in interplay community maps between PSKR1-SERK1 and RXEG1-BAK1 interactions (Fig. 5a, b). Moreover, the C3 area of BRI1 participates in each BR binding50,52 and co-receptor binding, while the C3 area of PSKR1 and RXEG1 solely engages in co-receptor interactions (SERK1/2 and BAK1, respectively)48,53. Through aligning the C3 areas of more than a few LRR-RLKs, together with BRI1/BRL-orthologs, PSKR orthologs, PSY1R, and a couple of LRR-RLPs, overlapping residues which are required for SERK interactions inside of PSKR/PSY1R, and LRR-RLP clades can also be discerned (See Effects segment ‘Specialisation of cell-surface receptors in numerous organic processes’ for alignment). For instance, the glutamic acid e residue at the second one place of the penultimate LRR motif is concerned about each PSKR1-SERK1 and RXEG1-BAK1 interactions53. This E residue is extremely conserved in each clades (PSKR/PSY1R clade: 69.9%, LRR-RLP: 86%). In a similar fashion, the phenylalanine (F) residue on the final place of the penultimate LRR motif contributes to each PSKR1-SERK1 and RXEG1-BAK1 interactions48. This F residue is conserved within the BRI1/BRL (62.2%), PSKR/PSY1R (99.7%), and LRR-RLP (64.1%) clades. There may be a conserved motif a very powerful for SERKs interactions throughout the final LRR motifs of BRI/BRL, PSKR/PSY1R, and LRR-RLP clades. This motif, Glutamine-x-x-Threonine/Serine (QxxT/S) loop, is conserved in BRI1/BRL (Q:86.6%; T/S:98.9%), PSKR/PSY1R (Q:99.9%; T/S:92.5%) and LRR-RLP (Q:91.8%; T/S:88.4%) clades, however it isn’t conserved in different LRR-RLPs out of doors of the ID + 4LRR clade or different LRR-RLK subgroups. Structural research of BRI1-BAK1, PSKR1-SERK1/2, and RXEG1-BAK1 additional helps the significance of residues inside of and across the QxxT/S loop for SERK interactions48,50,52,53 (Fig. 5a–c). In conclusion, our findings point out that the C3 areas of LRR-RLK-Xb and LRR-RLPs percentage a conserved serve as for interacting with SERKs.Fig. 5: Purposeful characterization of the C3 area in LRR-RLPs and LRR-RLK-Xb.
a Distribution of LRR-RLKs and LRR-RLPs in LRR-containing mobile floor receptors (LRR-TM) from 350 plant species, and the distribution of LRR-RLPs and LRR-RLKs without or with gaps of 10-29 amino acids (10-29) or 30-90 (30-90) amino acids. b, c Place of huge gaps (IDs; 30–90 AA) in b LRR-RLKs and c LRR-RLPs with a unmarried huge ID. N1 represents the choice of LRR motifs prior to the IDs and N2 represents the choice of LRR motifs after the ID. For positions of gaps in LRR-RLKs and LRR-RLPs with a couple of gaps, check with Supplementary Fig. 13. d The concentric ring pie chart gifts the proportion of LRR-containing cell-surface receptors (PRRs) from 350 species. The internal ring represents all LRR-containing cell-surface receptors (113,794); the center ring represents LRR-containing PRRs with ID (20,556); the outer ring represents LRR-containing PRRs with an ID previous the final 4 LRR (ID + 4LRR) on the C terminus (16,885). e The presence or absence of receptor categories in more than a few algal and plant lineages. *M/C/Ok represents Mesostigmatophyceae, Chlorokybophyceae, and Klebsormidiophyceae. A gray field signifies the absence, and a inexperienced field signifies the presence of a given receptor elegance in each and every lineage. The starting place of LRR-RLP and LRR-RLK-Xb with ID + 4LRR is indicated with a circle (○). f Series similarity tree of the C3 area (final 4 LRRs) from all LRR-containing cell-surface receptors of 350 species. Branches are colour-labelled as indicated. The internal ring and heart ring point out the lineage and subclass/order of the corresponding protein (species) from the department. Outer ring represents the LRR-RLP or LRR-RLK classification, which is indicated in d. The sunshine gray house signifies clustering of LRR-RLK-Xb and LRR-RLP with ID + 4LRR. The pruned collection similarity tree at the proper g corresponds to the sunshine gray house within the left tree, with clades labelled in darkish gray spaces accordingly. Characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade and the PSKR/PSY1R-clades also are labelled.Research of the distribution of LRR-RLK subgroups with IDs indicated that over 55% belong to the Xb subgroup (Fig. 4d). Moreover, 94.7% of LRR-RLKs with IDs located prior to the final 4 LRRs (ID + 4LRR) belong to the Xb subgroup (Fig. 4d), suggesting that each LRR-RLK-Xb and LRR-RLPs percentage the ID + 4LRR motif. A few of the characterized participants of LRR-RLK-Xb are essential elements of enlargement and advancement law, together with the BRASSINOSTEROID INSENSITIVE 1 (BRI1) members of the family, PHYTOSULFOKIN RECEPTOR 1 (PSKR1) and PSY1 RECEPTOR (PSY1R) members of the family, EXCESS MICROSPOROCYTES1 (EMS1) and NEMATODE-INDUCED LRR-RLK 1 (NILR1/GRACE)41,42,43,44,45. Curiously, we seen that each LRR-RLPs and LRR-RLK-Xbs with ID + 4LRR motifs are found in land crops (Embryophyte) however no longer in different lineages (Fig. 4e). Taking into account the similarity in structural motifs between LRR-RLPs and LRR-RLK-Xbs, it’s most probably that those two receptor categories percentage a not unusual starting place.To check this speculation, we carried out phylogenetic and structural analyses at the ectodomains of LRR-RLPs and LRR-RLKs. First, we aligned the final 4 LRR motifs (known as the C3 region46) from LRR-RLKs and LRR-RLPs (60,240). LRR-RLPs and LRR-RLK-Xbs with an ID + 4LRR cluster in combination within the C3 collection similarity tree, indicating a not unusual starting place (Fig. 4f). Inside this cluster (indicated because the ID + 4LRR clade), we seen distinct subclades, together with the BRI1/BRI1-LIKE (BRL) clade, the PSKR/PSY1R clade, and most commonly LRR-RLPs (Fig. 4g). The carefully equivalent C3 areas point out a conserved serve as. BRI1 recognises brassinosteroids (BRs) and interacts with the co-receptor BAK1 to urge BR responses47,48. In a similar fashion, PSKR1 perceives phytosulfokine (PSK) and interacts with the co-receptor SERK1/2. LRR-RLPs, then again, understand PAMPs or apoplastic effectors and interact each co-receptor BAK1 and SOBIR1 to start up immune responses49. Structural research have elucidated the interplay mechanism of BRI1, PSKR1, and the LRR-RLP RXEG1 with SERKs48,50,51,52,53,54 (Fig. 5a–c, Supplementary Fig. 14). Curiously, the C3 areas of BRI1, PSKR1, and RXEG1 play a a very powerful position in SERK interactions48,50,52,53, differentiating them from different LRR-RLKs. BRI1, PSKR1 and RXEG1 comprise particular amino acid residues throughout the terminal two LRR motifs (throughout the C3 area) to have interaction with BAK1/SERK1 (Fig. 5a–c), whilst the flagellin peptide (flg22) receptor FLS2 (an LRR-RLK-XII), is determined by the third–twelfth final LRR for BAK1 (SERK3) interactions55 (Supplementary Fig. 14a). The N-terminal area (or N-terminal cap) of BAK1 or SERK1 is essential for FLS2, BRI1, PSKR1, and RXEG1 interactions, whilst the first LRR internal floor of BAK1 or SERK1 is basically concerned about BRI1, PSKR1, and RXEG1 interactions, and the second and the 4th LRR internal floor of BAK1 is concerned about FLS2 interactions (Supplementary Fig. 14b). There also are putting similarities in interplay community maps between PSKR1-SERK1 and RXEG1-BAK1 interactions (Fig. 5a, b). Moreover, the C3 area of BRI1 participates in each BR binding50,52 and co-receptor binding, while the C3 area of PSKR1 and RXEG1 solely engages in co-receptor interactions (SERK1/2 and BAK1, respectively)48,53. Through aligning the C3 areas of more than a few LRR-RLKs, together with BRI1/BRL-orthologs, PSKR orthologs, PSY1R, and a couple of LRR-RLPs, overlapping residues which are required for SERK interactions inside of PSKR/PSY1R, and LRR-RLP clades can also be discerned (See Effects segment ‘Specialisation of cell-surface receptors in numerous organic processes’ for alignment). For instance, the glutamic acid e residue at the second one place of the penultimate LRR motif is concerned about each PSKR1-SERK1 and RXEG1-BAK1 interactions53. This E residue is extremely conserved in each clades (PSKR/PSY1R clade: 69.9%, LRR-RLP: 86%). In a similar fashion, the phenylalanine (F) residue on the final place of the penultimate LRR motif contributes to each PSKR1-SERK1 and RXEG1-BAK1 interactions48. This F residue is conserved within the BRI1/BRL (62.2%), PSKR/PSY1R (99.7%), and LRR-RLP (64.1%) clades. There may be a conserved motif a very powerful for SERKs interactions throughout the final LRR motifs of BRI/BRL, PSKR/PSY1R, and LRR-RLP clades. This motif, Glutamine-x-x-Threonine/Serine (QxxT/S) loop, is conserved in BRI1/BRL (Q:86.6%; T/S:98.9%), PSKR/PSY1R (Q:99.9%; T/S:92.5%) and LRR-RLP (Q:91.8%; T/S:88.4%) clades, however it isn’t conserved in different LRR-RLPs out of doors of the ID + 4LRR clade or different LRR-RLK subgroups. Structural research of BRI1-BAK1, PSKR1-SERK1/2, and RXEG1-BAK1 additional helps the significance of residues inside of and across the QxxT/S loop for SERK interactions48,50,52,53 (Fig. 5a–c). In conclusion, our findings point out that the C3 areas of LRR-RLK-Xb and LRR-RLPs percentage a conserved serve as for interacting with SERKs.Fig. 5: Purposeful characterization of the C3 area in LRR-RLPs and LRR-RLK-Xb.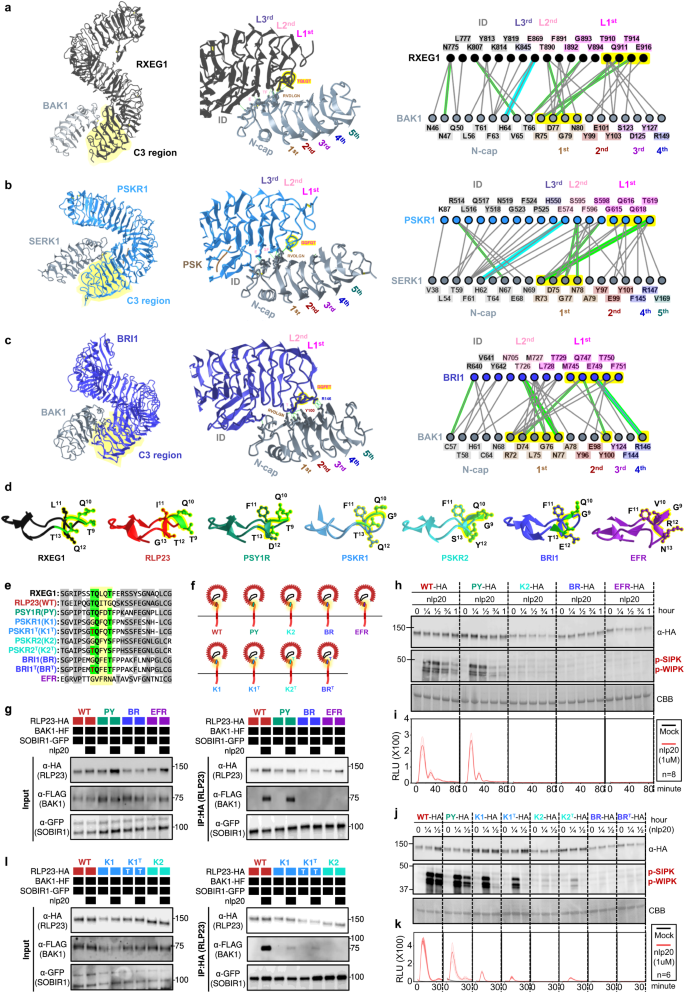 a–c Constructions and interplay interfaces of LRR-RLKs and LRR-RLPs with SERKs. Revealed buildings of a NbRXEG1-NbBAK153, b AtPSKR1-AtSERK148, and c AtBRI1-AtBAK150 and are proven. The left panels display the total construction, and the center panels display the interplay websites between LRR-RLKs or LRR-RLP and SERKs. Hydrogen bonds are indicated by way of inexperienced dotted strains, and salt bridges are proven as cyan dotted strains. The positions of LRR residues (counting from N to C for SERKs and counting from C to N for LRR-RLKs and LRR-RLP) are proven. Amino acid residues which are essential for the interactions are labelled and the QxxT motifs are highlighted in yellow (crimson textual content). The fitting panel represents the 2D interplay community between SERKs and the receptors. Contacts/interactions are proven in gray strains, hydrogen bonds are proven in inexperienced strains, and salt bridges are proven in cyan strains. Amino acids are labelled in colors in keeping with their positions within the LRR motifs (counting from N to C for SERKs and counting from C to N for LRR-RLKs and LRR-RLP l). Residues round and throughout the QxxT motifs in BRI1, PSKR1, and RXEG1 are highlighted in yellow. Residues in SERKs which are concerned within the interactions with QxxT motifs also are highlighted in yellow. Constructions had been visualized in iCn3D98. For a–c, the interplay websites are calculated by way of iCn3D with the next thresholds: hydrogen bonds: 4.2 Å; salt bridges/ionic bonds: 6 Å; contacts/interactions: 4 Å. d Construction of the terminal LRR motif of N. benthamiana (Nb)RXEG1, A. thaliana (At)RLP23, AtPSY1R, AtPSKR1, AtPSKR2, AtBRI1 and AtEFR. Constructions of NbRXEG1, AtPSKR1, and AtBRI1 had been published48,50,51,52,53,54. Constructions of AtRLP23, AtPSY1R, AtPSKR2 and AtEFR had been predicted by way of Alphafold297. Ectodomains are visualised in iCn3D98. e Alignment of amino acids within the final LRR motifs from NbRXEG1, AtRLP23, AtPSY1R, AtPSKR1, AtPSKR1T (G > T), AtPSKR2, AtPSKR2T (G > T), AtBRI1, AtBRI1T (G > T), and AtEFR. Amino acid residues concerned within the interplay between NbRXEG1 and BAK1 are highlighted in inexperienced. The QxxT motif positions are highlighted in yellow. Amino acids with equivalent houses to AtRLP23 are highlighted in gray. f Design of AtRLP23 chimeras. The final LRR motif of AtRLP23 is exchanged with both AtPSY1R, AtPSKR1, AtPSKR2, AtBRI1, or AtEFR. The glycine g residues in AtPSKR1, AtPSKR2, AtBRI1 have additionally been mutated to threonine (T). g, l Immuno-precipitation to check interactions between AtRLP23 chimeras, AtBAK1 and AtSOBIR1. Nb leaves expressing the indicated constructs had been handled with both mock or 1 μM nlp20 for five min. h–ok Capability checking out of AtRLP23 chimeras. Nb leaves expressing the indicated constructs had been handled with 1 μM nlp20 and samples had been gathered at indicated time issues. Phosphorylation of NbSIPK and NbWIPK used to be detected with p-P42/44 antibody. i, ok Nb leaf discs expressing the indicated constructs had been gathered and handled with both mock or 1 μM nlp20, and ROS manufacturing used to be measured for indicated time issues. For i and ok, cast line, imply; shaded band, s.e.m. RLU, relative gentle devices. For main points of experiential design in g–l, check with the strategies segment. For g, h, j and l, the experiments had been repeated no less than two times with equivalent effects.To additional validate the purposeful conservation of C3 areas in LRR-RLK-Xb and LRR-RLPs, we carried out purposeful research of the QxxT/S motif within the LRR-RLP RLP23 from A. thaliana. RLP23 paperwork heteromeric complexes with the LRR-RLK co-receptor SOBIR1, and upon the belief of the nlp20 peptide, BAK1 is recruited into the complicated, resulting in activation of the SOBIR1 KD to urge immunity20,49,56. Very similar to RXEG1 (with TQLQT), RLP23 possesses a TQITG motif (TQxxx), whilst LRR-RLK-Xb participants, similar to PSY1R, PSKR1, PSKR2, and BRI1, characteristic TQFDT, GQFQT, GQFYS, and GQFET motifs, respectively (all QxxT/S). Particularly, the LRR-RLK-XII member EFR lacks the QxxT/S motif in that place, having GVFRN as an alternative (Fig. 5d, e). We generated chimeric constructs of RLP23 with the terminal LRR motifs swapped between PSY1R, PSKR2, BRI1, and EFR (Fig. 5e, f). Through immuno-precipitation assays, we examined the power of those chimeras to have interaction with BAK1 upon ligand belief. Each wildtype RLP23 (WT) and RLP23-PSY1R (PY) chimeras can have interaction with BAK1 upon nlp20 remedy, while RLP23-BRI1 (BR) and RLP23-EFR (EFR) can’t (Fig. 5g). This means that the terminal LRR motif of RLP23 and PSY1R bind BAK1 in a equivalent method. All chimeras can have interaction with SOBIR1 irrespective of the presence of nlp20, indicating that the final LRR motif might not be concerned about SOBIR1 interactions (Fig. 5g). Moreover, upon nlp20 remedy, each WT and PY can cause immune responses, whilst K2 and BR can’t (Fig. 5h, i). We speculate that this can be because of the absence of a selected T residue prior to the Qxxx motif in PSKR2 and BRI1 (G as an alternative of T), which is fairly much less prevalent throughout the LRR-RLP clade (31.1%). A couple of A. thaliana LRR-RLPs comprise this sediment, however it isn’t in different studied LRR-RLPs, such because the tomato Cf proteins, suggesting that this sediment developed in some species after the divergence of LRR-RLK-Xb and LRR-RLPs. To check this speculation, we generated chimeric constructs of RLP23 with the terminal LRR motifs swapped to PSKR1 (K1), PSKR2 (K2), BRI1 (BR), and mutated the G residue into T (thereby K1T, K2T, BRT; Fig. 5e, f). We examined the immune responses precipitated by way of those chimeras following nlp20 remedy. Each K1, K1T, and K2T induce fairly immune responses in comparison to WT and PY, whilst K2, BR, and BRT are not able to urge any immune responses (Fig. 5j, ok). Each K1 and K1T weakly have interaction with BAK1 upon nlp20 remedy, whilst K2 does no longer have interaction with BAK1 (Fig. 5k). Thus, the residues round and throughout the TQxxx motif are each a very powerful for SERK interactions. We concluded that the C3 area in LRR-RLPs and a few LRR-RLK-Xbs (similar to PSY1R and PSKR1) have interaction with SERKs in a equivalent method, whilst some LRR-RLK-Xbs (PSKR2 and BRI1) have developed to interacts with SERKs in a rather other method. However, our effects strongly fortify the purposeful conservation of C3 areas in LRR-RLK-Xbs and LRR-RLPs, in particular their skill to have interaction with SERKs.At the starting place of LRR-RLPs and LRR-RLK-XbsThe purposeful conservation of C3 areas in LRR-RLK-Xbs and LRR-RLPs signifies that the ectodomains of those two receptor households may percentage a not unusual starting place. To dissect the ectodomain starting place of LRR-RLPs and LRR-RLK-Xbs, we investigated the relatedness of the IDs between those two teams of receptors. First, we aligned the IDs extracted from each LRR-RLKs and LRR-RLPs (20,246). Remarkably, the ID clusters of LRR-RLPs and LRR-RLK-Xb are present in shut proximity, mirroring the C3 collection similarity tree (Fig. 6a). Once more, the BRI1/BRI1-LIKE (BRL) and PSKR/PSY1R clusters are in proximity to the LRR-RLPs (Fig. 6a, b). Those effects also are in step with a prior record that PSKRs are carefully connected to a few LRR-RLPs in Arabidopsis and rice46. To dissect the ectodomain starting place of LRR-RLPs and LRR-RLK-Xbs. A up to date evaluation recognized two conserved lysine k-containing motifs, Yx8KG and Kx5Y, within the ID of LRR-RLPs40. The lysine residue within the Kx5Y motif from RXEG1 is needed for its interplay with BAK153. We recognized each Yx8KG and Kx5Y motifs within the extracted IDs (prior to the final 4LRR motifs) of LRR-RLKs and LRR-RLPs. Greater than 75% of IDs from LRR-RLPs comprise no less than this kind of lysine motifs, whilst not up to 5% of IDs from LRR-RLK-Xb have both motif (Fig. 6c). That is in step with structural information indicating that IDs from BRI1 and PSKR1 make use of distinct residues for his or her interactions with BAK148,50,52. Alternatively, IDs from LRR-RLPs which are carefully connected to these from LRR-RLK-Xbs retain Kx5Y motifs (Fig. 5b). It’s subsequently imaginable that the average ID ancestor can have firstly harboured lysine motifs that had been due to this fact misplaced from the BRI1 and PSKR/PSY1R clades.Fig. 6: At the starting place of LRR-RLPs and LRR-RLK-Xbs.
a–c Constructions and interplay interfaces of LRR-RLKs and LRR-RLPs with SERKs. Revealed buildings of a NbRXEG1-NbBAK153, b AtPSKR1-AtSERK148, and c AtBRI1-AtBAK150 and are proven. The left panels display the total construction, and the center panels display the interplay websites between LRR-RLKs or LRR-RLP and SERKs. Hydrogen bonds are indicated by way of inexperienced dotted strains, and salt bridges are proven as cyan dotted strains. The positions of LRR residues (counting from N to C for SERKs and counting from C to N for LRR-RLKs and LRR-RLP) are proven. Amino acid residues which are essential for the interactions are labelled and the QxxT motifs are highlighted in yellow (crimson textual content). The fitting panel represents the 2D interplay community between SERKs and the receptors. Contacts/interactions are proven in gray strains, hydrogen bonds are proven in inexperienced strains, and salt bridges are proven in cyan strains. Amino acids are labelled in colors in keeping with their positions within the LRR motifs (counting from N to C for SERKs and counting from C to N for LRR-RLKs and LRR-RLP l). Residues round and throughout the QxxT motifs in BRI1, PSKR1, and RXEG1 are highlighted in yellow. Residues in SERKs which are concerned within the interactions with QxxT motifs also are highlighted in yellow. Constructions had been visualized in iCn3D98. For a–c, the interplay websites are calculated by way of iCn3D with the next thresholds: hydrogen bonds: 4.2 Å; salt bridges/ionic bonds: 6 Å; contacts/interactions: 4 Å. d Construction of the terminal LRR motif of N. benthamiana (Nb)RXEG1, A. thaliana (At)RLP23, AtPSY1R, AtPSKR1, AtPSKR2, AtBRI1 and AtEFR. Constructions of NbRXEG1, AtPSKR1, and AtBRI1 had been published48,50,51,52,53,54. Constructions of AtRLP23, AtPSY1R, AtPSKR2 and AtEFR had been predicted by way of Alphafold297. Ectodomains are visualised in iCn3D98. e Alignment of amino acids within the final LRR motifs from NbRXEG1, AtRLP23, AtPSY1R, AtPSKR1, AtPSKR1T (G > T), AtPSKR2, AtPSKR2T (G > T), AtBRI1, AtBRI1T (G > T), and AtEFR. Amino acid residues concerned within the interplay between NbRXEG1 and BAK1 are highlighted in inexperienced. The QxxT motif positions are highlighted in yellow. Amino acids with equivalent houses to AtRLP23 are highlighted in gray. f Design of AtRLP23 chimeras. The final LRR motif of AtRLP23 is exchanged with both AtPSY1R, AtPSKR1, AtPSKR2, AtBRI1, or AtEFR. The glycine g residues in AtPSKR1, AtPSKR2, AtBRI1 have additionally been mutated to threonine (T). g, l Immuno-precipitation to check interactions between AtRLP23 chimeras, AtBAK1 and AtSOBIR1. Nb leaves expressing the indicated constructs had been handled with both mock or 1 μM nlp20 for five min. h–ok Capability checking out of AtRLP23 chimeras. Nb leaves expressing the indicated constructs had been handled with 1 μM nlp20 and samples had been gathered at indicated time issues. Phosphorylation of NbSIPK and NbWIPK used to be detected with p-P42/44 antibody. i, ok Nb leaf discs expressing the indicated constructs had been gathered and handled with both mock or 1 μM nlp20, and ROS manufacturing used to be measured for indicated time issues. For i and ok, cast line, imply; shaded band, s.e.m. RLU, relative gentle devices. For main points of experiential design in g–l, check with the strategies segment. For g, h, j and l, the experiments had been repeated no less than two times with equivalent effects.To additional validate the purposeful conservation of C3 areas in LRR-RLK-Xb and LRR-RLPs, we carried out purposeful research of the QxxT/S motif within the LRR-RLP RLP23 from A. thaliana. RLP23 paperwork heteromeric complexes with the LRR-RLK co-receptor SOBIR1, and upon the belief of the nlp20 peptide, BAK1 is recruited into the complicated, resulting in activation of the SOBIR1 KD to urge immunity20,49,56. Very similar to RXEG1 (with TQLQT), RLP23 possesses a TQITG motif (TQxxx), whilst LRR-RLK-Xb participants, similar to PSY1R, PSKR1, PSKR2, and BRI1, characteristic TQFDT, GQFQT, GQFYS, and GQFET motifs, respectively (all QxxT/S). Particularly, the LRR-RLK-XII member EFR lacks the QxxT/S motif in that place, having GVFRN as an alternative (Fig. 5d, e). We generated chimeric constructs of RLP23 with the terminal LRR motifs swapped between PSY1R, PSKR2, BRI1, and EFR (Fig. 5e, f). Through immuno-precipitation assays, we examined the power of those chimeras to have interaction with BAK1 upon ligand belief. Each wildtype RLP23 (WT) and RLP23-PSY1R (PY) chimeras can have interaction with BAK1 upon nlp20 remedy, while RLP23-BRI1 (BR) and RLP23-EFR (EFR) can’t (Fig. 5g). This means that the terminal LRR motif of RLP23 and PSY1R bind BAK1 in a equivalent method. All chimeras can have interaction with SOBIR1 irrespective of the presence of nlp20, indicating that the final LRR motif might not be concerned about SOBIR1 interactions (Fig. 5g). Moreover, upon nlp20 remedy, each WT and PY can cause immune responses, whilst K2 and BR can’t (Fig. 5h, i). We speculate that this can be because of the absence of a selected T residue prior to the Qxxx motif in PSKR2 and BRI1 (G as an alternative of T), which is fairly much less prevalent throughout the LRR-RLP clade (31.1%). A couple of A. thaliana LRR-RLPs comprise this sediment, however it isn’t in different studied LRR-RLPs, such because the tomato Cf proteins, suggesting that this sediment developed in some species after the divergence of LRR-RLK-Xb and LRR-RLPs. To check this speculation, we generated chimeric constructs of RLP23 with the terminal LRR motifs swapped to PSKR1 (K1), PSKR2 (K2), BRI1 (BR), and mutated the G residue into T (thereby K1T, K2T, BRT; Fig. 5e, f). We examined the immune responses precipitated by way of those chimeras following nlp20 remedy. Each K1, K1T, and K2T induce fairly immune responses in comparison to WT and PY, whilst K2, BR, and BRT are not able to urge any immune responses (Fig. 5j, ok). Each K1 and K1T weakly have interaction with BAK1 upon nlp20 remedy, whilst K2 does no longer have interaction with BAK1 (Fig. 5k). Thus, the residues round and throughout the TQxxx motif are each a very powerful for SERK interactions. We concluded that the C3 area in LRR-RLPs and a few LRR-RLK-Xbs (similar to PSY1R and PSKR1) have interaction with SERKs in a equivalent method, whilst some LRR-RLK-Xbs (PSKR2 and BRI1) have developed to interacts with SERKs in a rather other method. However, our effects strongly fortify the purposeful conservation of C3 areas in LRR-RLK-Xbs and LRR-RLPs, in particular their skill to have interaction with SERKs.At the starting place of LRR-RLPs and LRR-RLK-XbsThe purposeful conservation of C3 areas in LRR-RLK-Xbs and LRR-RLPs signifies that the ectodomains of those two receptor households may percentage a not unusual starting place. To dissect the ectodomain starting place of LRR-RLPs and LRR-RLK-Xbs, we investigated the relatedness of the IDs between those two teams of receptors. First, we aligned the IDs extracted from each LRR-RLKs and LRR-RLPs (20,246). Remarkably, the ID clusters of LRR-RLPs and LRR-RLK-Xb are present in shut proximity, mirroring the C3 collection similarity tree (Fig. 6a). Once more, the BRI1/BRI1-LIKE (BRL) and PSKR/PSY1R clusters are in proximity to the LRR-RLPs (Fig. 6a, b). Those effects also are in step with a prior record that PSKRs are carefully connected to a few LRR-RLPs in Arabidopsis and rice46. To dissect the ectodomain starting place of LRR-RLPs and LRR-RLK-Xbs. A up to date evaluation recognized two conserved lysine k-containing motifs, Yx8KG and Kx5Y, within the ID of LRR-RLPs40. The lysine residue within the Kx5Y motif from RXEG1 is needed for its interplay with BAK153. We recognized each Yx8KG and Kx5Y motifs within the extracted IDs (prior to the final 4LRR motifs) of LRR-RLKs and LRR-RLPs. Greater than 75% of IDs from LRR-RLPs comprise no less than this kind of lysine motifs, whilst not up to 5% of IDs from LRR-RLK-Xb have both motif (Fig. 6c). That is in step with structural information indicating that IDs from BRI1 and PSKR1 make use of distinct residues for his or her interactions with BAK148,50,52. Alternatively, IDs from LRR-RLPs which are carefully connected to these from LRR-RLK-Xbs retain Kx5Y motifs (Fig. 5b). It’s subsequently imaginable that the average ID ancestor can have firstly harboured lysine motifs that had been due to this fact misplaced from the BRI1 and PSKR/PSY1R clades.Fig. 6: At the starting place of LRR-RLPs and LRR-RLK-Xbs.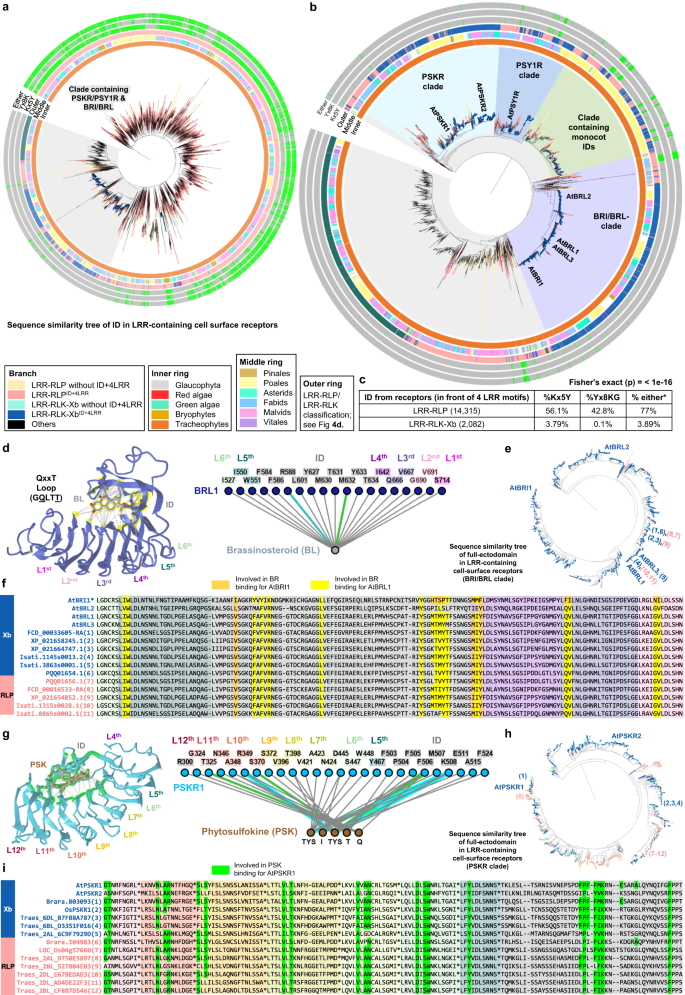 a Series similarity tree of IDs extracted from all LRR-containing PRRs. Branches are labelled in colors as indicated. Gray clade represents clade that comprises each PSKR/PSY1R & BRI/BRL circle of relatives, this clade is proven in b. In b, characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade, the PSKR clade, the PSY1R clade, and a clade with monocot-only IDs are labelled in numerous colors. In a, b, internal ring (Internal), heart ring (Center) and outer ring (Outer) are labelled as indicated. Outer ring represents LRR-RLP and LRR-RLKs classification proven in Fig. 4d. Within the outmost 3 rings, the presence of Kx5Y (Kx5Y), Yx8K (Yx8K), or both Kx5Y or Yx8K (Both) within the IDs are indicated in inexperienced, and lack of those motifs is indicated in gray. c Proportion (%) of IDs (prior to the final 4 LRR motifs) from LRR-RLP and LRR-RLK-Xb with the Kx5Y, Yx8KG, or both Kx5Y or Yx8KG (*) motifs. The Fisher check (2-sided) used to be carried out to match the quantity/fraction of IDs with both Kx5Y or Yx8KG in LRR-RLPs towards LRR-RLK-Xb. The calculated p-value mentioned right here (<1e-16) is simply too low to be given actual quantity. Thus, the higher sure restrict is mentioned as an alternative. d, g Constructions and interplay interfaces of AtBRI1-Brassinosteroid (BR) and AtPSKR1-Phytosulfokine (PSK). Revealed buildings of d AtBRL1-BR100, g AtPSKR1-PSK48 are proven. The left panels display the interplay websites between the LRR-RLK-Xb receptors and their ligands. Contacts are indicated by way of gray strains, hydrogen bonds are indicated by way of inexperienced dotted strains, and salt bridges are proven as cyan dotted strains. The positions of LRR residues (counting from C to N) are proven. Amino acid residues which are essential for the interactions are highlighted in yellow and inexperienced, respectively. The fitting panel represents the 2D interplay community between the LRR-RLK-Xb receptors and their ligands. Contacts/interactions are proven in gray strains, hydrogen bonds are proven in inexperienced strains, and salt bridges are proven in cyan strains. Amino acids are labelled in colors in keeping with their positions within the LRR motifs (counting from C to N). Constructions had been visualized in iCn3D98. e, h Series similarity timber of the full-ectodomains of e BRI1/BRL clade and h PSKR clade from 350 species. Branches are labelled in colors as indicated in a. Those timber are extracted from the BRI1/BRL and PSKR branches from Supplementary Fig. 16. In e, h, characterised LRR-RLK-Xb participants are labelled. The LRR-RLK-Xb and LRR-RLP participants taken for the alignment in f and that i also are labelled in blue numbers (LRR-RLK-Xb) and red numbers (LRR-RLP), respectively. f, i Ectodomain and alignment of a couple of LRR-RLK-Xb and LRR-RLP participants throughout the BRI1/BRL-clade extracted from e and the PSKR-clade extracted from h. f The alignment of ectodomain from LRR-RLK-Xb (blue) and LRR-RLP (red) participants taken from the collection similarity tree in e. The orange highlights point out the amino acids residues required from BR binding in AtBRI1, and the yellow highlights point out the amino acid residues required for BR binding in AtBRL1100. The yellow highlights are akin to the amino acids highlighted within the construction in d. The LRR motifs and ID within the alignment are indicated in colors proven in d (the interplay community; proper panel). i The alignment of ectodomain from LRR-RLK-Xb (blue) and LRR-RLP (red) participants taken from the collection similarity tree in h. The fairway highlights point out the amino acids residues required from PSK binding in AtPSKR148. The fairway highlights are akin to the amino acids highlighted within the construction in g. The LRR motifs and ID within the alignment are indicated in colors proven in g (the interplay community; proper panel). Because of area limitation, the final 9 amino acids in each and every LRR motif are introduced as * within the alignment. For the total alignment, check with Supplementary Fig. 17a.To additional discover the relatedness of the IDs between LRR-RLK-Xb team and LRR-RLPs, we shaped clusters of extremely equivalent IDs and tested their subgroup/circle of relatives affiliations. Total, we recognized greater than 2,822 clusters, with the bulk (2,734) consisting of LRR-receptors of a unmarried subgroup/circle of relatives (Supplementary Fig. 15). A few of the 61 clusters containing LRR-receptors from two other subgroups/households, 80% (49 clusters) include LRR-receptors from LRR-RLK-Xb and LRR-RLPs (Supplementary Fig. 15). The enrichment of LRR-RLK-Xb and LRR-RLP pairings supplies additional proof for the relatedness in their ectodomains. The really extensive choice of LRR-RLP-only ID clusters (2,383) additionally means that the IDs of LRR-RLPs have gone through in depth evolution and diversification, within the procedure offering a wide scope for the popularity of PAMPs and apoplastic effectors. Maximum clusters comprise fairly a small choice of IDs, predominantly from species inside of the similar order or circle of relatives (Supplementary Information 2). Because of this, tracing again the starting place of IDs is difficult because of their substantial range. We subsequently suggest that the IDs of LRR-RLK-Xbs and LRR-RLPs most probably originated from a not unusual ancestor, with the IDs of LRR-RLPs increasing and diversifying after the divergence of LRR-RLK-Xb and LRR-RLPs.Throughout the BRI1/BRI1-LIKE (BRL) and PSKR/PSY1R clusters, we discovered LRR-RLP opposite numbers of BRI1, PSKR, and PSY1R. To additional check their relatedness, we aligned the total ectodomains from LRR-RLKs and LRR-RLPs and extracted the clades containing BRI1, PSKR, and PSY1R from the collection similarity tree (Supplementary Fig. 16a, b). Throughout the BRI1/BRL- and PSKR-ectodomain clades (Fig. 6e, h), we discovered ectodomains from LRR-RLPs which are extremely very similar to BRL1/BRL3 and PSKR2, with residues very important for BR and PSK binding, respectively48,54 (Fig. 6d–l). Throughout the PSY1R-ectodomain subclade, we discovered a couple of IDs from LRR-RLPs that percentage outstanding similarity with AtPSY1R (Supplementary Fig. 17b, c). AtRLP3 and AtPSY1R have over 70% collection identification and 85% similarity of their ectodomains. Despite the fact that PSY1R isn’t the receptor for PSY peptide57, it’s imaginable that RLP3, and PSY1R recognise equivalent or equivalent ligands58. These days, the purposes of those BRL-, PSKR- and PSY1R-like LRR-RLPs stay unclear. AtRLP3 confers resistance towards the vascular wilt fungus Fusarium oxysporum f. sp. matthioli58, whilst AtPSY1R is concerned about enlargement and development43. We recommend that those RLPs might both recognise endogenous molecules to turn on enlargement and advancement, or take part within the popularity of pathogen-mimicking molecules to cause immune signalling.Specialisation of cell-surface receptors in numerous organic processesGiven the average starting place of the ectodomains of LRR-RLPs and LRR-RLK-Xbs, those receptors should have gone through specialisation in immune- and developmental processes following their divergence. Whilst LRR-RLK-Xbs turn on downstream responses during the Xb kinase area, LRR-RLPs recruit SOBIR1 to cause immunity49,56. As a result of LRR-RLPs lack a kinase area, the juxtamembrane (JM) and TM areas might turn on immune responses. We subsequently aligned the C3, eJM (JM area prior to TM), TM, and cJM (JM area after TM, however with the absence of kinase area from RLKs) areas from LRR-RLKs and LRR-RLPs (with a subset of 40, or the total set of 62,896 cell-surface receptors; Fig. 7a–c). Curiously, the eJM-TM-cJM area of LRR-RLPs isn’t carefully associated with LRR-RLK-Xbs (Fig. 7c). Earlier research have reported the requirement of Glycine-X-X-X-Glycine (GxxxG) motifs in SOBIR1 for its affiliation with LRR-RLPs, and the possible contribution of negatively charged amino acids within the eJM area of LRR-RLPs to their interplay with SOBIR159. In keeping with earlier reports59, LRR-RLPs are strongly negatively charged on the finish of the eJM area, while LRR-RLKs, together with SOBIR1, are definitely charged on the finish of the eJM area (Fig. 7a). Maximum LRR-RLPs have a unmarried GxxxG motif, with some having two (GxxxGxxxG) or 3 (GxxxGxxxGxxxG) consecutive motifs (Fig. 7a). Conversely, GxxxG motifs are fairly much less not unusual in LRR-RLKs, however can also be present in SOBIR1, BAK1, and the PSKR/PSY1R clade (Fig. 7a). We additional tested general fees within the eJM area of the LRR-RLP and LRR-RLK households. Maximum LRR-RLK subgroups characteristic definitely charged eJM areas, while LRR-RLPs have negatively charged eJM areas. Importantly, negatively charged eJM areas are solely found in LRR-RLPs throughout the ID + 4LRR clade (Fig. 7d). Greater than 80% of LRR-RLPs throughout the ID + 4LRR clade comprise GxxxG motifs of their TMs. Additionally, GxxxGxxxG motifs are basically present in LRR-RLPs and are fairly uncommon in LRR-RLKs (Fig. 4d). Curiously, each GxxxG and GxxxGxxxG motifs are fairly enriched within the PSKR/PSY1R clade, indicating that the TM areas of LRR-RLPs and PSYR/PSY1R clade may also percentage a not unusual starting place. (Fig. 7d).Fig. 7: Alignment and contours of the terminal 4 C3, eJM, and TM area in LRR-RLKs and LRR-RLPs.
a Series similarity tree of IDs extracted from all LRR-containing PRRs. Branches are labelled in colors as indicated. Gray clade represents clade that comprises each PSKR/PSY1R & BRI/BRL circle of relatives, this clade is proven in b. In b, characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade, the PSKR clade, the PSY1R clade, and a clade with monocot-only IDs are labelled in numerous colors. In a, b, internal ring (Internal), heart ring (Center) and outer ring (Outer) are labelled as indicated. Outer ring represents LRR-RLP and LRR-RLKs classification proven in Fig. 4d. Within the outmost 3 rings, the presence of Kx5Y (Kx5Y), Yx8K (Yx8K), or both Kx5Y or Yx8K (Both) within the IDs are indicated in inexperienced, and lack of those motifs is indicated in gray. c Proportion (%) of IDs (prior to the final 4 LRR motifs) from LRR-RLP and LRR-RLK-Xb with the Kx5Y, Yx8KG, or both Kx5Y or Yx8KG (*) motifs. The Fisher check (2-sided) used to be carried out to match the quantity/fraction of IDs with both Kx5Y or Yx8KG in LRR-RLPs towards LRR-RLK-Xb. The calculated p-value mentioned right here (<1e-16) is simply too low to be given actual quantity. Thus, the higher sure restrict is mentioned as an alternative. d, g Constructions and interplay interfaces of AtBRI1-Brassinosteroid (BR) and AtPSKR1-Phytosulfokine (PSK). Revealed buildings of d AtBRL1-BR100, g AtPSKR1-PSK48 are proven. The left panels display the interplay websites between the LRR-RLK-Xb receptors and their ligands. Contacts are indicated by way of gray strains, hydrogen bonds are indicated by way of inexperienced dotted strains, and salt bridges are proven as cyan dotted strains. The positions of LRR residues (counting from C to N) are proven. Amino acid residues which are essential for the interactions are highlighted in yellow and inexperienced, respectively. The fitting panel represents the 2D interplay community between the LRR-RLK-Xb receptors and their ligands. Contacts/interactions are proven in gray strains, hydrogen bonds are proven in inexperienced strains, and salt bridges are proven in cyan strains. Amino acids are labelled in colors in keeping with their positions within the LRR motifs (counting from C to N). Constructions had been visualized in iCn3D98. e, h Series similarity timber of the full-ectodomains of e BRI1/BRL clade and h PSKR clade from 350 species. Branches are labelled in colors as indicated in a. Those timber are extracted from the BRI1/BRL and PSKR branches from Supplementary Fig. 16. In e, h, characterised LRR-RLK-Xb participants are labelled. The LRR-RLK-Xb and LRR-RLP participants taken for the alignment in f and that i also are labelled in blue numbers (LRR-RLK-Xb) and red numbers (LRR-RLP), respectively. f, i Ectodomain and alignment of a couple of LRR-RLK-Xb and LRR-RLP participants throughout the BRI1/BRL-clade extracted from e and the PSKR-clade extracted from h. f The alignment of ectodomain from LRR-RLK-Xb (blue) and LRR-RLP (red) participants taken from the collection similarity tree in e. The orange highlights point out the amino acids residues required from BR binding in AtBRI1, and the yellow highlights point out the amino acid residues required for BR binding in AtBRL1100. The yellow highlights are akin to the amino acids highlighted within the construction in d. The LRR motifs and ID within the alignment are indicated in colors proven in d (the interplay community; proper panel). i The alignment of ectodomain from LRR-RLK-Xb (blue) and LRR-RLP (red) participants taken from the collection similarity tree in h. The fairway highlights point out the amino acids residues required from PSK binding in AtPSKR148. The fairway highlights are akin to the amino acids highlighted within the construction in g. The LRR motifs and ID within the alignment are indicated in colors proven in g (the interplay community; proper panel). Because of area limitation, the final 9 amino acids in each and every LRR motif are introduced as * within the alignment. For the total alignment, check with Supplementary Fig. 17a.To additional discover the relatedness of the IDs between LRR-RLK-Xb team and LRR-RLPs, we shaped clusters of extremely equivalent IDs and tested their subgroup/circle of relatives affiliations. Total, we recognized greater than 2,822 clusters, with the bulk (2,734) consisting of LRR-receptors of a unmarried subgroup/circle of relatives (Supplementary Fig. 15). A few of the 61 clusters containing LRR-receptors from two other subgroups/households, 80% (49 clusters) include LRR-receptors from LRR-RLK-Xb and LRR-RLPs (Supplementary Fig. 15). The enrichment of LRR-RLK-Xb and LRR-RLP pairings supplies additional proof for the relatedness in their ectodomains. The really extensive choice of LRR-RLP-only ID clusters (2,383) additionally means that the IDs of LRR-RLPs have gone through in depth evolution and diversification, within the procedure offering a wide scope for the popularity of PAMPs and apoplastic effectors. Maximum clusters comprise fairly a small choice of IDs, predominantly from species inside of the similar order or circle of relatives (Supplementary Information 2). Because of this, tracing again the starting place of IDs is difficult because of their substantial range. We subsequently suggest that the IDs of LRR-RLK-Xbs and LRR-RLPs most probably originated from a not unusual ancestor, with the IDs of LRR-RLPs increasing and diversifying after the divergence of LRR-RLK-Xb and LRR-RLPs.Throughout the BRI1/BRI1-LIKE (BRL) and PSKR/PSY1R clusters, we discovered LRR-RLP opposite numbers of BRI1, PSKR, and PSY1R. To additional check their relatedness, we aligned the total ectodomains from LRR-RLKs and LRR-RLPs and extracted the clades containing BRI1, PSKR, and PSY1R from the collection similarity tree (Supplementary Fig. 16a, b). Throughout the BRI1/BRL- and PSKR-ectodomain clades (Fig. 6e, h), we discovered ectodomains from LRR-RLPs which are extremely very similar to BRL1/BRL3 and PSKR2, with residues very important for BR and PSK binding, respectively48,54 (Fig. 6d–l). Throughout the PSY1R-ectodomain subclade, we discovered a couple of IDs from LRR-RLPs that percentage outstanding similarity with AtPSY1R (Supplementary Fig. 17b, c). AtRLP3 and AtPSY1R have over 70% collection identification and 85% similarity of their ectodomains. Despite the fact that PSY1R isn’t the receptor for PSY peptide57, it’s imaginable that RLP3, and PSY1R recognise equivalent or equivalent ligands58. These days, the purposes of those BRL-, PSKR- and PSY1R-like LRR-RLPs stay unclear. AtRLP3 confers resistance towards the vascular wilt fungus Fusarium oxysporum f. sp. matthioli58, whilst AtPSY1R is concerned about enlargement and development43. We recommend that those RLPs might both recognise endogenous molecules to turn on enlargement and advancement, or take part within the popularity of pathogen-mimicking molecules to cause immune signalling.Specialisation of cell-surface receptors in numerous organic processesGiven the average starting place of the ectodomains of LRR-RLPs and LRR-RLK-Xbs, those receptors should have gone through specialisation in immune- and developmental processes following their divergence. Whilst LRR-RLK-Xbs turn on downstream responses during the Xb kinase area, LRR-RLPs recruit SOBIR1 to cause immunity49,56. As a result of LRR-RLPs lack a kinase area, the juxtamembrane (JM) and TM areas might turn on immune responses. We subsequently aligned the C3, eJM (JM area prior to TM), TM, and cJM (JM area after TM, however with the absence of kinase area from RLKs) areas from LRR-RLKs and LRR-RLPs (with a subset of 40, or the total set of 62,896 cell-surface receptors; Fig. 7a–c). Curiously, the eJM-TM-cJM area of LRR-RLPs isn’t carefully associated with LRR-RLK-Xbs (Fig. 7c). Earlier research have reported the requirement of Glycine-X-X-X-Glycine (GxxxG) motifs in SOBIR1 for its affiliation with LRR-RLPs, and the possible contribution of negatively charged amino acids within the eJM area of LRR-RLPs to their interplay with SOBIR159. In keeping with earlier reports59, LRR-RLPs are strongly negatively charged on the finish of the eJM area, while LRR-RLKs, together with SOBIR1, are definitely charged on the finish of the eJM area (Fig. 7a). Maximum LRR-RLPs have a unmarried GxxxG motif, with some having two (GxxxGxxxG) or 3 (GxxxGxxxGxxxG) consecutive motifs (Fig. 7a). Conversely, GxxxG motifs are fairly much less not unusual in LRR-RLKs, however can also be present in SOBIR1, BAK1, and the PSKR/PSY1R clade (Fig. 7a). We additional tested general fees within the eJM area of the LRR-RLP and LRR-RLK households. Maximum LRR-RLK subgroups characteristic definitely charged eJM areas, while LRR-RLPs have negatively charged eJM areas. Importantly, negatively charged eJM areas are solely found in LRR-RLPs throughout the ID + 4LRR clade (Fig. 7d). Greater than 80% of LRR-RLPs throughout the ID + 4LRR clade comprise GxxxG motifs of their TMs. Additionally, GxxxGxxxG motifs are basically present in LRR-RLPs and are fairly uncommon in LRR-RLKs (Fig. 4d). Curiously, each GxxxG and GxxxGxxxG motifs are fairly enriched within the PSKR/PSY1R clade, indicating that the TM areas of LRR-RLPs and PSYR/PSY1R clade may also percentage a not unusual starting place. (Fig. 7d).Fig. 7: Alignment and contours of the terminal 4 C3, eJM, and TM area in LRR-RLKs and LRR-RLPs.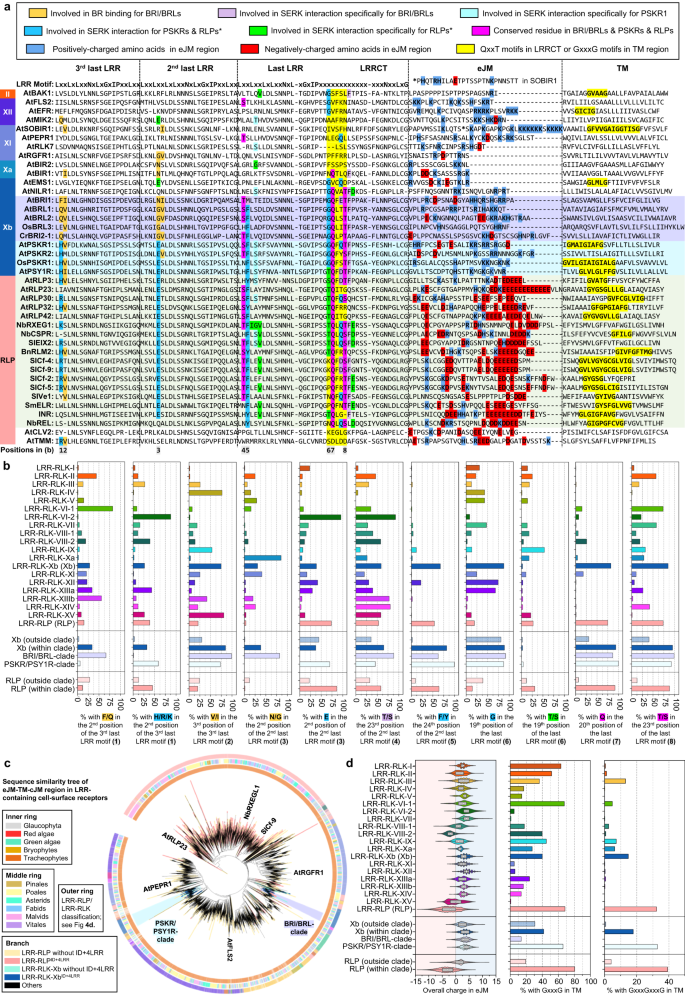 a Alignment of the C3, eJM, and TM area in LRR-RLKs (from subgroups II, XII, XI, Xa, Xb) and LRR-RLPs. The BRI1/BRL clade is highlighted in crimson; PSKR/PSY1R clade is highlighted in cyan and LRR-RLP with the ID + 4LRR clade (see Fig. 4c) is highlighted in gentle inexperienced. Amino acid residues concerned about brassinosteroid (BR) binding for BRI/BRL and residues required for SERK interplay for BRI1/BRL, PSKR1, and RXEG1 (LRR-RLP) are highlighted. The color code for each and every spotlight is indicated within the field on most sensible. For the eJM area, amino acid residues with damaging fees are highlighted in crimson, and amino acids with certain fees are in highlighted blue. The GxxxG motif within the TM area is highlighted in yellow. The interplay websites had been calculated the usage of iCn3D98 with the next thresholds: hydrogen bonds: 4.2 Å; salt bridges/ionic bonds: 6 Å; contacts/interactions: 4 Å. For main points, please refer Fig. 5. b Percentages of LRR-RLKs or LRR-RLPs with the mentioned amino acid residues within the corresponding place in a. Percentages (%) had been calculated by way of the choice of LRR-RLKs or LRR-RLPs within the subgroup with the mentioned residue divided by way of the choice of LRR-RLKs or LRR-RLPs within the subgroup with out the mentioned residue × 100. c Series similarity tree of the eJM-TM-cJM area from all LRR-containing cell-surface receptors of 350 species. Branches are colour-labelled as indicated. The internal ring and heart ring point out the lineage and subclass/order of the corresponding protein (species) from the department. Outer ring represents the LRR-RLP or LRR-RLK classification, which is indicated in Fig. 4d. Characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade and the PSKR/PSY1R-clades also are labelled. d Total fee distribution in eJM (left), proportion of receptors with GxxxG (heart), and GxxxGxxxG (proper) within the TM area. For b, d RLP/RLK-Xb (out of doors clade) refers to receptors out of doors the sunshine gray clade in Fig. 4f. RLP/RLK-Xb (inside of clade) refers to receptors within the gentle gray clade in Fig. 4f. Collection of cell-surface receptors (n) in each and every LRR-RLK subgroup: I, n = 752; II, n = 682; III, n = 6572; IV, n = 1033; V, n = 8; VI-1, n = 84; V1-2, n = 146; VII, n = 1720; VIII-1, n = 195; VIII-2, n = 411; IX, n = 70; Xa, n = 96; Xb, n = 3182; XI, n = 8807; XII, n = 12863; XIIIa, n = 739; XIIIb, n = 465; XIV, n = 241; XV, n = 548; Xb (out of doors clade), n = 580; Xb (inside of clade), n = 2527; BRI/BRL clade, n = 1170; PSKR/PSY1R clade, n = 1347. Collection of cell-surface receptors (n) in each and every LRR-RLP subgroup: LRR-RLP (RLP), n = 24970; RLP (out of doors clade), n = 5000; RLP (inside of clade), n = 19970. Boxplot components: middle line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles.To evaluate the capability of eJM-TM-cytosolic areas in LRR-RLK-Xbs and LRR-RLPs, we generated a couple of chimeras of eJM, TM, and cytosolic areas of BRI1 and RLP23 (Fig. 8a). In RLP23c-BRI1, the cytosolic area (following TM) of BRI1 used to be swapped into RLP23. In RLP23TM-BRI1, the TM+ cytosolic area of BRI1 used to be swapped into RLP23. In RLP23eJM-BRI1, the eJM + TM + cytosolic area of BRI1 used to be swapped into RLP23 (Fig. 8a). Immuno-precipitation assays confirmed that each RLP23WT and RLP23c-BRI1 showcase constitutive interactions with SOBIR1, and in particular have interaction with BAK1 upon nlp20 remedy. RLP23TM-BRI1 does no longer have interaction with SOBIR1 however nonetheless interacts with BAK1 upon nlp20 remedy, suggesting that the RLP23 TM area with GxxxG motifs is vital for SOBIR1-, however no longer BAK1-interactions (Fig. 8b). In keeping with those immunoprecipitation assays, RLP23WT and RLP23c-BRI1 can turn on immune responses, whilst RLP23c-BRI1 and RLP23TM-BRI1 can turn on developmental responses as indicated by way of the dephosphorylation of BRI1-EMS-SUPPRESSOR 1 (BES1)60 (Fig. 8c; Supplementary Fig. 18). This confirms that the BRI1 kinase area is in particular required for the activation of BR responses. Curiously, RLP23TM-BRI1 too can turn on susceptible immune responses, most probably unbiased of SOBIR1 (Fig. 8c, d). RLP23eJM-BRI1 does no longer have interaction with SOBIR1 or BAK1, and is not able to turn on both immune or developmental responses (Fig. 8c, d). Those information counsel that the eJM area of RLP23 is vital for RLP23-BAK1 interactions. This conclusion used to be bolstered by way of producing RLP23eo-BRI1 chimera, by which the eJM area of RLP23 is changed with the BRI1 eJM (Fig. 8a). RLP23eo-BRI1 persistently accumulates much less protein than RLP23WT. Additionally, RLP23eo-BRI1 constitutively interacts with SOBIR1, however fails to have interaction with BAK1 with nlp20 remedy (Fig. 8e). RLP23eo-BRI1 solely weakly turns on MAPKs and fails to cause ROS manufacturing with nlp20 remedy (Fig. 8f, g). Thus, we conclude that the eJM areas of LRR-RLPs are essential for protein accumulation and interplay with BAK1 upon ligand belief. The eJM, TM, and cytosolic area of LRR-RLPs and LRR-RLK-Xbs have certainly specialised to recruit other proteins into the receptor complicated, which permits them to turn on differential downstream responses (Fig. 8h).Fig. 8: Adaptation of LRRID + 4LRR to differential downstream signalling pathways.
a Alignment of the C3, eJM, and TM area in LRR-RLKs (from subgroups II, XII, XI, Xa, Xb) and LRR-RLPs. The BRI1/BRL clade is highlighted in crimson; PSKR/PSY1R clade is highlighted in cyan and LRR-RLP with the ID + 4LRR clade (see Fig. 4c) is highlighted in gentle inexperienced. Amino acid residues concerned about brassinosteroid (BR) binding for BRI/BRL and residues required for SERK interplay for BRI1/BRL, PSKR1, and RXEG1 (LRR-RLP) are highlighted. The color code for each and every spotlight is indicated within the field on most sensible. For the eJM area, amino acid residues with damaging fees are highlighted in crimson, and amino acids with certain fees are in highlighted blue. The GxxxG motif within the TM area is highlighted in yellow. The interplay websites had been calculated the usage of iCn3D98 with the next thresholds: hydrogen bonds: 4.2 Å; salt bridges/ionic bonds: 6 Å; contacts/interactions: 4 Å. For main points, please refer Fig. 5. b Percentages of LRR-RLKs or LRR-RLPs with the mentioned amino acid residues within the corresponding place in a. Percentages (%) had been calculated by way of the choice of LRR-RLKs or LRR-RLPs within the subgroup with the mentioned residue divided by way of the choice of LRR-RLKs or LRR-RLPs within the subgroup with out the mentioned residue × 100. c Series similarity tree of the eJM-TM-cJM area from all LRR-containing cell-surface receptors of 350 species. Branches are colour-labelled as indicated. The internal ring and heart ring point out the lineage and subclass/order of the corresponding protein (species) from the department. Outer ring represents the LRR-RLP or LRR-RLK classification, which is indicated in Fig. 4d. Characterised LRR-RLK-Xb and LRR-RLP participants are labelled. The BRI/BRL-clade and the PSKR/PSY1R-clades also are labelled. d Total fee distribution in eJM (left), proportion of receptors with GxxxG (heart), and GxxxGxxxG (proper) within the TM area. For b, d RLP/RLK-Xb (out of doors clade) refers to receptors out of doors the sunshine gray clade in Fig. 4f. RLP/RLK-Xb (inside of clade) refers to receptors within the gentle gray clade in Fig. 4f. Collection of cell-surface receptors (n) in each and every LRR-RLK subgroup: I, n = 752; II, n = 682; III, n = 6572; IV, n = 1033; V, n = 8; VI-1, n = 84; V1-2, n = 146; VII, n = 1720; VIII-1, n = 195; VIII-2, n = 411; IX, n = 70; Xa, n = 96; Xb, n = 3182; XI, n = 8807; XII, n = 12863; XIIIa, n = 739; XIIIb, n = 465; XIV, n = 241; XV, n = 548; Xb (out of doors clade), n = 580; Xb (inside of clade), n = 2527; BRI/BRL clade, n = 1170; PSKR/PSY1R clade, n = 1347. Collection of cell-surface receptors (n) in each and every LRR-RLP subgroup: LRR-RLP (RLP), n = 24970; RLP (out of doors clade), n = 5000; RLP (inside of clade), n = 19970. Boxplot components: middle line, median; bounds of field, twenty fifth and seventy fifth percentiles; whiskers, 1.5 × IQR from twenty fifth and seventy fifth percentiles.To evaluate the capability of eJM-TM-cytosolic areas in LRR-RLK-Xbs and LRR-RLPs, we generated a couple of chimeras of eJM, TM, and cytosolic areas of BRI1 and RLP23 (Fig. 8a). In RLP23c-BRI1, the cytosolic area (following TM) of BRI1 used to be swapped into RLP23. In RLP23TM-BRI1, the TM+ cytosolic area of BRI1 used to be swapped into RLP23. In RLP23eJM-BRI1, the eJM + TM + cytosolic area of BRI1 used to be swapped into RLP23 (Fig. 8a). Immuno-precipitation assays confirmed that each RLP23WT and RLP23c-BRI1 showcase constitutive interactions with SOBIR1, and in particular have interaction with BAK1 upon nlp20 remedy. RLP23TM-BRI1 does no longer have interaction with SOBIR1 however nonetheless interacts with BAK1 upon nlp20 remedy, suggesting that the RLP23 TM area with GxxxG motifs is vital for SOBIR1-, however no longer BAK1-interactions (Fig. 8b). In keeping with those immunoprecipitation assays, RLP23WT and RLP23c-BRI1 can turn on immune responses, whilst RLP23c-BRI1 and RLP23TM-BRI1 can turn on developmental responses as indicated by way of the dephosphorylation of BRI1-EMS-SUPPRESSOR 1 (BES1)60 (Fig. 8c; Supplementary Fig. 18). This confirms that the BRI1 kinase area is in particular required for the activation of BR responses. Curiously, RLP23TM-BRI1 too can turn on susceptible immune responses, most probably unbiased of SOBIR1 (Fig. 8c, d). RLP23eJM-BRI1 does no longer have interaction with SOBIR1 or BAK1, and is not able to turn on both immune or developmental responses (Fig. 8c, d). Those information counsel that the eJM area of RLP23 is vital for RLP23-BAK1 interactions. This conclusion used to be bolstered by way of producing RLP23eo-BRI1 chimera, by which the eJM area of RLP23 is changed with the BRI1 eJM (Fig. 8a). RLP23eo-BRI1 persistently accumulates much less protein than RLP23WT. Additionally, RLP23eo-BRI1 constitutively interacts with SOBIR1, however fails to have interaction with BAK1 with nlp20 remedy (Fig. 8e). RLP23eo-BRI1 solely weakly turns on MAPKs and fails to cause ROS manufacturing with nlp20 remedy (Fig. 8f, g). Thus, we conclude that the eJM areas of LRR-RLPs are essential for protein accumulation and interplay with BAK1 upon ligand belief. The eJM, TM, and cytosolic area of LRR-RLPs and LRR-RLK-Xbs have certainly specialised to recruit other proteins into the receptor complicated, which permits them to turn on differential downstream responses (Fig. 8h).Fig. 8: Adaptation of LRRID + 4LRR to differential downstream signalling pathways. a Design of AtRLP23-BRI1 chimeras. Other areas (cytosolic, TM+cytosolic, and eJM+TM+cytosolic) of BRI1 had been swapped into AtRLP23 as indicated. The alignment of amino acids within the eJM and TM areas from AtRLP23 and AtBRI1 is proven. Amino acid residues with damaging fees are in crimson and amino acids with certain fees are in blue. The GxxxG motif within the TM area is highlighted in yellow. b, e Immuno-precipitation to check interactions between AtRLP23 chimeras, AtBAK1 and AtSOBIR1. Nb leaves expressing the indicated constructs had been handled with both mock or 1 μM nlp20 for five min. c, d, f, g Capability checking out of AtRLP23 chimeras. Nb leaves expressing the indicated constructs had been handled with 1 μM nlp20 and samples had been gathered at indicated time issues. Dephosphorylation of BESI1-HA used to be detected with HA antibody. Phosphorylation of NbSIPK and NbWIPK used to be detected with p-P42/44 antibody. For f, two times the pattern of RLP23oe-BRI1 used to be loaded as a reference, as a result of RLP23oe-BRI1 protein accumulation is weaker than that of RLP23WT. d, g Nb leaf discs expressing the indicated constructs (with out BES1-HA) had been gathered and handled with both mock or 1 μM nlp20 and ROS manufacturing used to be measured for 90 min. For d and g, cast line, imply; shaded band, s.e.m. RLU, relative gentle devices. For main points of experiential design, check with the strategies segment. h Schematic style of the interplay between LRR-RLK-XbID+4LRR and LRR-RLPID + 4LRR with co-receptors to urge differential downstream signalling. Each receptor categories utilise the final 4 LRRs (highlighted in yellow) to have interaction with SERKs (BAK1). LRR-RLP developed to have interaction with SOBIR1 with the GxxxG motifs in TM (highlighted in yellow define). Colored hexagons on RLKs point out activated kinases and black hexagon signifies an inactivated kinase. i Modular evolution of various domain names in cell-surface receptors to permit various ligand popularity and specificity of downstream signalling. Domain names or areas that developed other purposes are highlighted in yellow. Daring arrows constitute huge expansions and variations. Ok* represents the lysine in Kx5Y or Yx8KG motifs in ID from LRR-RLPs. Area or area buildings (from left to proper) are bought from: BRI1 ectodomain (3RGX); RXEG1 ectodomain (7W3X); PSKR1 ID (4Z63); RXEG1 ID (7W3X); PSKR1 C3 (4Z63); RXEG1 C3 (7W3X); PSKR1 ejM-TM (predicted from Alphafold220); RLP23 eJM-TM (predicted from Alphafold220); BRI1 kinase (4OH4). Constructions had been visualized in iCn3D98. For b, c, e and f, the experiments had been repeated no less than two times with equivalent effects.
a Design of AtRLP23-BRI1 chimeras. Other areas (cytosolic, TM+cytosolic, and eJM+TM+cytosolic) of BRI1 had been swapped into AtRLP23 as indicated. The alignment of amino acids within the eJM and TM areas from AtRLP23 and AtBRI1 is proven. Amino acid residues with damaging fees are in crimson and amino acids with certain fees are in blue. The GxxxG motif within the TM area is highlighted in yellow. b, e Immuno-precipitation to check interactions between AtRLP23 chimeras, AtBAK1 and AtSOBIR1. Nb leaves expressing the indicated constructs had been handled with both mock or 1 μM nlp20 for five min. c, d, f, g Capability checking out of AtRLP23 chimeras. Nb leaves expressing the indicated constructs had been handled with 1 μM nlp20 and samples had been gathered at indicated time issues. Dephosphorylation of BESI1-HA used to be detected with HA antibody. Phosphorylation of NbSIPK and NbWIPK used to be detected with p-P42/44 antibody. For f, two times the pattern of RLP23oe-BRI1 used to be loaded as a reference, as a result of RLP23oe-BRI1 protein accumulation is weaker than that of RLP23WT. d, g Nb leaf discs expressing the indicated constructs (with out BES1-HA) had been gathered and handled with both mock or 1 μM nlp20 and ROS manufacturing used to be measured for 90 min. For d and g, cast line, imply; shaded band, s.e.m. RLU, relative gentle devices. For main points of experiential design, check with the strategies segment. h Schematic style of the interplay between LRR-RLK-XbID+4LRR and LRR-RLPID + 4LRR with co-receptors to urge differential downstream signalling. Each receptor categories utilise the final 4 LRRs (highlighted in yellow) to have interaction with SERKs (BAK1). LRR-RLP developed to have interaction with SOBIR1 with the GxxxG motifs in TM (highlighted in yellow define). Colored hexagons on RLKs point out activated kinases and black hexagon signifies an inactivated kinase. i Modular evolution of various domain names in cell-surface receptors to permit various ligand popularity and specificity of downstream signalling. Domain names or areas that developed other purposes are highlighted in yellow. Daring arrows constitute huge expansions and variations. Ok* represents the lysine in Kx5Y or Yx8KG motifs in ID from LRR-RLPs. Area or area buildings (from left to proper) are bought from: BRI1 ectodomain (3RGX); RXEG1 ectodomain (7W3X); PSKR1 ID (4Z63); RXEG1 ID (7W3X); PSKR1 C3 (4Z63); RXEG1 C3 (7W3X); PSKR1 ejM-TM (predicted from Alphafold220); RLP23 eJM-TM (predicted from Alphafold220); BRI1 kinase (4OH4). Constructions had been visualized in iCn3D98. For b, c, e and f, the experiments had been repeated no less than two times with equivalent effects.
Evolutionary trajectory of trend popularity receptors in crops – Nature Communications