# New E. coli strain will accelerate evolution of the genes of your choice
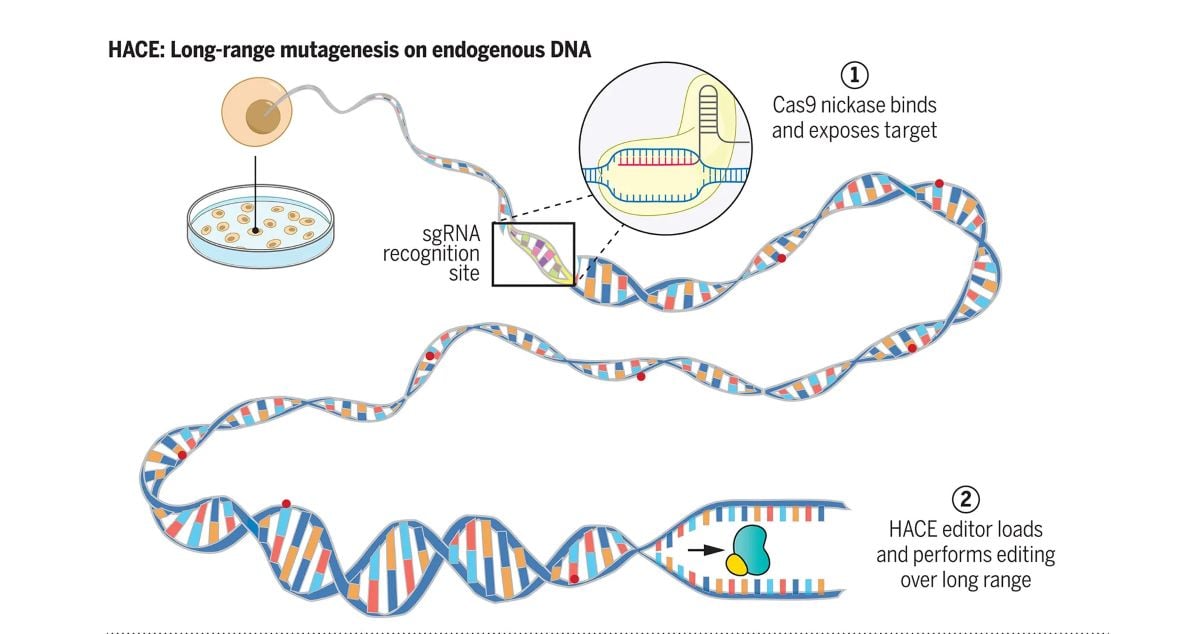
Genetic mutations are crucial for innovation and evolution. However, too many or the wrong kind of mutations can be detrimental. To address this challenge, researchers at Cambridge have developed a synthetic “orthogonal” DNA replication system in E. coli. This system provides a low-risk method for generating and studying mutations. The orthologous system is completely separate from the mechanism that E. coli uses to replicate its genome, which contains the essential genes for the bacterium’s survival.
The genes in the orthogonal system are copied using an extremely error-prone DNA replication enzyme, which accelerates evolution by producing numerous random mutations. This occurs while E. coli’s essential genes are replicated by its normal high-fidelity DNA copying enzyme. Both enzymes work simultaneously without interfering with each other’s functions.
The concept of this system was inspired by nature, as yeast already possesses a similar system with a set of genes copied by a dedicated enzyme that does not replicate the rest of the genome. However, E. coli is easier to work with than yeast and its population can double in just 20 minutes, allowing for rapid rounds of replication and evolution.
The researchers created the system by modifying a phage, a virus that infects E. coli, removing all of the genes that facilitate uncontrolled growth until the phage bursts the infected E. coli cell. This engineering left behind a cassette containing the genes responsible for copying the phage genome. When this cassette was inserted into the E. coli genome, it could concurrently replicate at least three different sets of genes placed next to it in the DNA, while leaving the rest of the E. coli genome to be copied by other enzymes.
The scientists adjusted the mutation rate of the orthogonal DNA-replicating enzyme, ultimately increasing it by 1,000-fold. To test the system’s ability to evolve new functions, they inserted a gene for resistance to one antibiotic and observed how long it took for that gene to mutate into one providing resistance to a different antibiotic. Within twelve days, they achieved 150 times more resistance to the new antibiotic. They also inserted the gene encoding green fluorescent protein and increased its fluorescence over 1,000-fold in just five days.
In the same issue of Science, Frances Arnold’s lab presented a study that demonstrates the effectiveness of this approach. This team directed the evolution of an enzyme using traditional methods: through sequential rounds of random mutagenesis and selection for the desired trait. Arnold, who received the Nobel Prize in Chemistry in 2018 for the directed evolution of enzymes, led this effort. In their recent work, her lab developed an enzyme capable of biodegrading volatile methyl siloxanes. These compounds are used in a variety of everyday products but persist in the environment due to the lack of a natural degradation process. This new orthologous system will hopefully greatly facilitate the development of enzymes and other proteins with applications in research, medicine, and industry.
While machines that can efficiently assemble long stretches of DNA or make proteins are currently unavailable, cells perform these tasks with great efficiency. E. coli cells have long been used as little factories in the lab to produce genes or proteins as needed. Now, E. coli can also serve as a hub for evolution.
**Science**, 2024. DOI: 10.1126/science.adi5554, 10.1126/science.adk1281