Affected person baseline demographics and specimen handlingOur Molecular Dual Pilot (MT-Pilot) cohort incorporated 74 sufferers at medical degree I (n = 47) and II (n = 27) with surgically resected PDAC between March 2015 and April 2019. Tumor specimens have been gathered on the time of surgical procedure and plasma specimens have been gathered preoperatively. DS used to be recorded and handled as a binary finish level on the time of study as of 21 October 2021. Right now, 45 (61%) sufferers have been deceased. All demographic and medical traits (Supplementary Desk 1) have been incorporated as options for the medical analyte in our multi-omic research. The surgical pathology data used to be acquired from a pancreas tumor resection. Tumor and plasma specimens have been assessed for person options by means of molecular profiling, together with focused next-generation DNA sequencing (NGS), full-transcriptome RNA sequencing, paired (tumor and standard from the similar affected person) tissue proteomics, unpaired (tumor from sufferers and standard unrelated controls) plasma proteomics, lipidomics and computational pathology. Analyte profiling yielded options that we used to validate single- and multi-omic fashions for predicting DS; a leave-one-out cross-validation method used to be carried out to the MT-Pilot cohort, while the 4 unbiased datasets: TCGA, JHU cohort 1, JHU cohort 2 and MGH have been used to validate our ML fashions and have panels advanced according to the MT-Pilot information (Fig. 1).Fig. 1: Find out about classification technique review.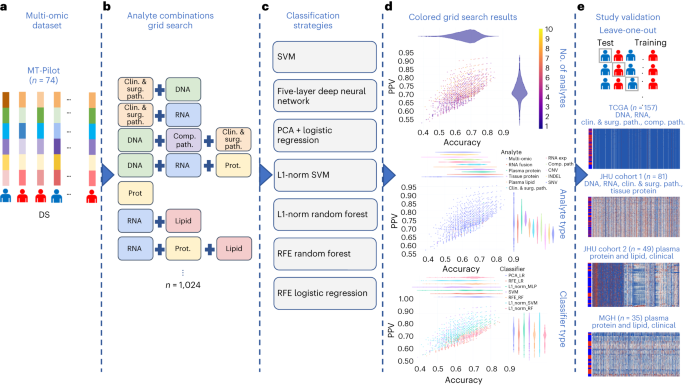 a, Mixed multi-omic dataset of 6,363 processed options spanning medical and surgical pathology, SNV, CNV, INDEL, RNA, fusion, tissue proteins, plasma proteins, lipids and computational pathology analytes. b, Building of all conceivable analyte mixtures (n = 1,024) by means of a drop-column significance method to simulate availability of quite a lot of mixtures of analytes. c, For each and every analyte aggregate, seven unbiased ML fashions have been skilled for fashion analysis, together with SVM, principal-component research (PCA) + logistic regression, L1-normalized SVM, L1-normalized RF, five-hidden-layer deep neural community, RFE logistic regression and RFE RF. d, Enter analyte mixtures (n = 1,024) with seven modeling methods in keeping with analyte aggregate produced 7,168 ensuing grid seek runs that have been due to this fact analyzed for predictive energy, analyte composition and have contributions for DS prediction. e, Every distinctive analyte aggregate and ML technique used to be skilled by means of leave-one-patient-out cross-validation method. Unmarried-omic and multi-omic fashions for DS prediction have been validated the use of checking out units from 4 separate cohorts, TCGA, JHU cohort 1, JHU cohort 2 and the MGH cohort. Clin. & surg. trail., medical and surgical pathology; comp. trail., computational pathology; prot., protein.Supply dataClinical and surgical pathology options affect outcomesThe 331 medical options, together with surgical pathology options and chemotherapy medicine (Supplementary Desk 1), in addition to comorbidities (Supplementary Desk 2) have been analyzed the use of more than one ML fashions. When skilled with those options, the random wooded area (RF) used to be the tip acting fashion in figuring out DS and accomplished an accuracy of 0.70 (95% self belief period (CI) 0.60–0.81) and sure predictive price (PPV) of 0.71 (95% CI 0.60–0.82) (Desk 1and Prolonged Information Fig. 1). The highest options predicting end result incorporated comorbidities, similar to hyperlipidemia, jaundice and pancreatitis, in addition to surgical margin standing (Supplementary Desk 2), which might be recognized within the PDAC field15,16,17. The fashion for DS used to be predominantly pushed by means of comorbid prerequisites, which accounted for 306 of the 331 overall options. The RF fashion used to be additionally skilled the use of the remainder 25 options, which incorporated recognized PDAC predictors similar to earlier chemotherapy and margin standing. This fashion carried out in a similar way to ones that incorporated all medical options (Supplementary Desk 2). Significantly, the tip ten options of this fashion incorporated surgical margin standing, tumor grade and chemotherapy, which might be recognized to persuade affected person outcome18,19.Desk 1 Best single-omic and multi-omic analytes for predicting illness survival in PDAC within the MT-Pilot cohortDNA research finds alterations with prognostic importancePoint mutations and insertion/deletion (INDEL) polymorphisms are not unusual in established PDAC oncogenes and tumor suppressor genes20. Tissue samples have been processed for 611 somatic single-nucleotide variants (SNVs), 648 copy-number diversifications (CNVs) and 126 INDELs. Those options have been then utilized in affected person DS prediction fashions (Supplementary Desk 3).The use of SNV options, the top-performing fashion to resolve DS used to be RF, with accuracy of 0.64 (95% CI 0.53–0.75) and PPV of 0.66 (95% CI 0.55–0.77) (Desk 1 and Prolonged Information Fig. 1). In fashions comparing SNVs, we discovered that alterations in RAD51, IL6R, FGF20 and SOX2 genes have been the tip DS predictors (Supplementary Desk 3) and their related signaling pathways have essential prognostic implications in PDAC21,22,23. As well as, we discovered genes, similar to RIT1, that have been height predictive DS markers recognized by means of our fashion and now not in the past related to PDAC diagnosis or targetable pathways.The use of CNV options, the top-performing fashion to resolve DS used to be an RF fashion with an accuracy of 0.65 (95% CI 0.57–0.80) and PPV of 0.68 (95% CI 0.57–0.80) (Desk 1and Prolonged Information Fig. 1). The highest CNV options for DS are famous in Supplementary Desk 3. Significantly, we discovered that FOXQ1 and KDM5D have been height predictors related to DS. Each are markers for PDAC diagnosis and possible healing targets24,25,26. In our cohort, the 4 frequently mutated genes, KRAS, TP53, CDKN2A and SMAD4 (ref. 27), have been incorporated amongst a complete of 126 particular INDEL options and have been realized by means of more than one ML fashion sorts. The highest fashion predicting DS used to be RF with an accuracy of 0.64 (95% CI 0.53–0.75) and PPV of 0.70 (95% CI 0.58–0.82) (Desk 1 and Prolonged Information Fig. 1). The highest options within the fashion incorporated mutations of TP53, CDKN2A and SMAD4 (refs. 28,29), which were proven to correlate with deficient diagnosis and extra competitive phenotypes of PDAC.RNA signatures of drug resistance have an effect on prognosisWhole-transcriptome sequencing used to be carried out on 72 of the 74 formalin-fixed paraffin-embedded (FFPE) tumor tissue samples. To optimize probably the most predictive options, we first ran a differential expression research between most cancers and noncancer samples from the GTex Consortium to choose RNA gene transcripts for downstream modeling30. The highest-performing fashion to resolve DS used to be L1-normalized RF, which yielded an accuracy of 0.68 (95% CI 0.56–0.80) and PPV of 0.70 (95% CI 0.57–0.83) (Desk 1 and Prolonged Information Fig. 1). In our height fashion for DS prediction the NFE2L2 and LRIG3 genes have been the 2 height options (Supplementary Desk 4). Contemporary investigations have proven that the NRF2 pathway via NFE2L2 regulates resistance to medication and immunotherapy31,32. Moreover, a complete of 29 RNA fusions have been analyzed the use of more than one fashion sorts (Supplementary Desk 4). The highest acting fashion that includes RNA fusions to resolve DS, used to be give a boost to vector gadget (SVM) with an accuracy of 0.75 (95% CI 0.64–0.87) and PPV of 0.74 (95% CI 0.62–0.87) (Desk 1 and Prolonged Information Fig. 1).Plasma proteins are crucial biomarkers in survival predictionProteomics and lipidomics research to begin with generated 3,777 tumor tissue proteomic, 1,051 plasma proteomic and 939 lipidomic options (Supplementary Desk 5).The use of tissue protein options, the tip acting fashion to expect DS used to be RF fashion with accuracy of 0.73 (95% CI 0.61–0.86) and PPV of 0.76 (95% CI 0.63–0.89) (Desk 1 and Prolonged Information Fig. 1). For plasma protein options, the top-performing fashion for DS used to be the five-hidden-layer-deep neural community fashion with an accuracy of 0.75 (95% CI 0.63–0.86) and PPV of 0.80 (95% CI 0.68–0.90) (Desk 1 and Prolonged Information Fig. 1). Amongst DS predictive plasma proteins, we recognized ANXA1, which is the most important rising participant in pancreatic carcinogenesis and PDAC drug resistance33,34. The highest acting fashion the use of plasma lipid options to resolve DS used to be the RF fashion with an accuracy of 0.71 (95% CI 0.58–0.83) and PPV of 0.74 (95% CI 0.61–0.87) (Desk 1 and Prolonged Information Fig. 1). The highest plasma lipidomics options for DS have been pushed by means of diacylglycerols and cholesteryl esters (Supplementary Desk 5).As mentioned above, CA 19-9 is mechanically used in medical observe at PDAC analysis, pre- and postoperatively to evaluate illness biology, medicine reaction and diagnosis. CA 19-9 readouts acquired at analysis, prior to surgical procedure and postoperatively, have been realized by means of the RF fashion, however the DS prediction had a low accuracy (0.59–0.64, 95% CI 0.47–0.76) and PPV (0.52–0.61, 95% CI 0.40–73) throughout all time issues (Supplementary Desk 6).Predictive nuclear morphology by means of computational pathologyThe 71 hematoxylin and eosin (H&E)-stained PDAC tissue whole-slide photographs (WSIs) have been evaluated by means of a synthetic intelligence (AI)-based computational pathology pipeline (Fig. 2). The pipeline incorporated two convolutional neural community fashions: a fashion to mask-out most cancers cells (Fig. 2a) and a fashion to delineate nuclei (Fig. 2b). When examined on photographs from an unbiased set of 40 PDAC circumstances, the cancer-masking fashion accomplished 0.90 world accuracy, 0.784 imply intersection over union (mIoU) and imply F1-scores of 0.83 and nil.77 in figuring out nontumor and tumor tissue pixels, respectively. Subsequent, the pipeline used to be run on 2,908 areas (~41 ± 11 areas in keeping with case) randomly decided on from the 71 WSIs in our cohort and routinely remoted 345,038 tumor mobile nuclei (~4,860 nuclei in keeping with case). Nuclear morphology and texture have been quantitated by means of a panel of 63 traits. Distribution of traits in each and every case used to be additional summarized by means of 13 order statistics, yielding 819 options in keeping with case (Fig. 2c and Supplementary Desk 7). A uniform manifold approximation and projection (UMAP) plot published clusters of circumstances with an identical end result (Fig. second) suggesting that one of the most options within the panel endure prognostic possible. The use of the leave-one-out method and the 819 options in keeping with case, we cross-validated seven classification fashions for DS prediction. An RF with an accuracy of 0.66 (95% CI 0.55–0.77) and PPV of 0.76 (95% CI 0.63–0.88) (Fig. 2e) carried out the most productive. All over all validation steps, options realized by means of the tip fashions have been ranked according to the have an effect on on predicting the result and the frequency of prevalence of impactful options measured. Impactful options that came about in a minimum of 10% of validation steps have been thought to be height options. The 17 out of 39 height options to expect survival in Fig. 2f originated from the similar 10 out of 63 nuclear traits in Fig. 2c.Fig. 2: Computational pathology pipeline.
a, Mixed multi-omic dataset of 6,363 processed options spanning medical and surgical pathology, SNV, CNV, INDEL, RNA, fusion, tissue proteins, plasma proteins, lipids and computational pathology analytes. b, Building of all conceivable analyte mixtures (n = 1,024) by means of a drop-column significance method to simulate availability of quite a lot of mixtures of analytes. c, For each and every analyte aggregate, seven unbiased ML fashions have been skilled for fashion analysis, together with SVM, principal-component research (PCA) + logistic regression, L1-normalized SVM, L1-normalized RF, five-hidden-layer deep neural community, RFE logistic regression and RFE RF. d, Enter analyte mixtures (n = 1,024) with seven modeling methods in keeping with analyte aggregate produced 7,168 ensuing grid seek runs that have been due to this fact analyzed for predictive energy, analyte composition and have contributions for DS prediction. e, Every distinctive analyte aggregate and ML technique used to be skilled by means of leave-one-patient-out cross-validation method. Unmarried-omic and multi-omic fashions for DS prediction have been validated the use of checking out units from 4 separate cohorts, TCGA, JHU cohort 1, JHU cohort 2 and the MGH cohort. Clin. & surg. trail., medical and surgical pathology; comp. trail., computational pathology; prot., protein.Supply dataClinical and surgical pathology options affect outcomesThe 331 medical options, together with surgical pathology options and chemotherapy medicine (Supplementary Desk 1), in addition to comorbidities (Supplementary Desk 2) have been analyzed the use of more than one ML fashions. When skilled with those options, the random wooded area (RF) used to be the tip acting fashion in figuring out DS and accomplished an accuracy of 0.70 (95% self belief period (CI) 0.60–0.81) and sure predictive price (PPV) of 0.71 (95% CI 0.60–0.82) (Desk 1and Prolonged Information Fig. 1). The highest options predicting end result incorporated comorbidities, similar to hyperlipidemia, jaundice and pancreatitis, in addition to surgical margin standing (Supplementary Desk 2), which might be recognized within the PDAC field15,16,17. The fashion for DS used to be predominantly pushed by means of comorbid prerequisites, which accounted for 306 of the 331 overall options. The RF fashion used to be additionally skilled the use of the remainder 25 options, which incorporated recognized PDAC predictors similar to earlier chemotherapy and margin standing. This fashion carried out in a similar way to ones that incorporated all medical options (Supplementary Desk 2). Significantly, the tip ten options of this fashion incorporated surgical margin standing, tumor grade and chemotherapy, which might be recognized to persuade affected person outcome18,19.Desk 1 Best single-omic and multi-omic analytes for predicting illness survival in PDAC within the MT-Pilot cohortDNA research finds alterations with prognostic importancePoint mutations and insertion/deletion (INDEL) polymorphisms are not unusual in established PDAC oncogenes and tumor suppressor genes20. Tissue samples have been processed for 611 somatic single-nucleotide variants (SNVs), 648 copy-number diversifications (CNVs) and 126 INDELs. Those options have been then utilized in affected person DS prediction fashions (Supplementary Desk 3).The use of SNV options, the top-performing fashion to resolve DS used to be RF, with accuracy of 0.64 (95% CI 0.53–0.75) and PPV of 0.66 (95% CI 0.55–0.77) (Desk 1 and Prolonged Information Fig. 1). In fashions comparing SNVs, we discovered that alterations in RAD51, IL6R, FGF20 and SOX2 genes have been the tip DS predictors (Supplementary Desk 3) and their related signaling pathways have essential prognostic implications in PDAC21,22,23. As well as, we discovered genes, similar to RIT1, that have been height predictive DS markers recognized by means of our fashion and now not in the past related to PDAC diagnosis or targetable pathways.The use of CNV options, the top-performing fashion to resolve DS used to be an RF fashion with an accuracy of 0.65 (95% CI 0.57–0.80) and PPV of 0.68 (95% CI 0.57–0.80) (Desk 1and Prolonged Information Fig. 1). The highest CNV options for DS are famous in Supplementary Desk 3. Significantly, we discovered that FOXQ1 and KDM5D have been height predictors related to DS. Each are markers for PDAC diagnosis and possible healing targets24,25,26. In our cohort, the 4 frequently mutated genes, KRAS, TP53, CDKN2A and SMAD4 (ref. 27), have been incorporated amongst a complete of 126 particular INDEL options and have been realized by means of more than one ML fashion sorts. The highest fashion predicting DS used to be RF with an accuracy of 0.64 (95% CI 0.53–0.75) and PPV of 0.70 (95% CI 0.58–0.82) (Desk 1 and Prolonged Information Fig. 1). The highest options within the fashion incorporated mutations of TP53, CDKN2A and SMAD4 (refs. 28,29), which were proven to correlate with deficient diagnosis and extra competitive phenotypes of PDAC.RNA signatures of drug resistance have an effect on prognosisWhole-transcriptome sequencing used to be carried out on 72 of the 74 formalin-fixed paraffin-embedded (FFPE) tumor tissue samples. To optimize probably the most predictive options, we first ran a differential expression research between most cancers and noncancer samples from the GTex Consortium to choose RNA gene transcripts for downstream modeling30. The highest-performing fashion to resolve DS used to be L1-normalized RF, which yielded an accuracy of 0.68 (95% CI 0.56–0.80) and PPV of 0.70 (95% CI 0.57–0.83) (Desk 1 and Prolonged Information Fig. 1). In our height fashion for DS prediction the NFE2L2 and LRIG3 genes have been the 2 height options (Supplementary Desk 4). Contemporary investigations have proven that the NRF2 pathway via NFE2L2 regulates resistance to medication and immunotherapy31,32. Moreover, a complete of 29 RNA fusions have been analyzed the use of more than one fashion sorts (Supplementary Desk 4). The highest acting fashion that includes RNA fusions to resolve DS, used to be give a boost to vector gadget (SVM) with an accuracy of 0.75 (95% CI 0.64–0.87) and PPV of 0.74 (95% CI 0.62–0.87) (Desk 1 and Prolonged Information Fig. 1).Plasma proteins are crucial biomarkers in survival predictionProteomics and lipidomics research to begin with generated 3,777 tumor tissue proteomic, 1,051 plasma proteomic and 939 lipidomic options (Supplementary Desk 5).The use of tissue protein options, the tip acting fashion to expect DS used to be RF fashion with accuracy of 0.73 (95% CI 0.61–0.86) and PPV of 0.76 (95% CI 0.63–0.89) (Desk 1 and Prolonged Information Fig. 1). For plasma protein options, the top-performing fashion for DS used to be the five-hidden-layer-deep neural community fashion with an accuracy of 0.75 (95% CI 0.63–0.86) and PPV of 0.80 (95% CI 0.68–0.90) (Desk 1 and Prolonged Information Fig. 1). Amongst DS predictive plasma proteins, we recognized ANXA1, which is the most important rising participant in pancreatic carcinogenesis and PDAC drug resistance33,34. The highest acting fashion the use of plasma lipid options to resolve DS used to be the RF fashion with an accuracy of 0.71 (95% CI 0.58–0.83) and PPV of 0.74 (95% CI 0.61–0.87) (Desk 1 and Prolonged Information Fig. 1). The highest plasma lipidomics options for DS have been pushed by means of diacylglycerols and cholesteryl esters (Supplementary Desk 5).As mentioned above, CA 19-9 is mechanically used in medical observe at PDAC analysis, pre- and postoperatively to evaluate illness biology, medicine reaction and diagnosis. CA 19-9 readouts acquired at analysis, prior to surgical procedure and postoperatively, have been realized by means of the RF fashion, however the DS prediction had a low accuracy (0.59–0.64, 95% CI 0.47–0.76) and PPV (0.52–0.61, 95% CI 0.40–73) throughout all time issues (Supplementary Desk 6).Predictive nuclear morphology by means of computational pathologyThe 71 hematoxylin and eosin (H&E)-stained PDAC tissue whole-slide photographs (WSIs) have been evaluated by means of a synthetic intelligence (AI)-based computational pathology pipeline (Fig. 2). The pipeline incorporated two convolutional neural community fashions: a fashion to mask-out most cancers cells (Fig. 2a) and a fashion to delineate nuclei (Fig. 2b). When examined on photographs from an unbiased set of 40 PDAC circumstances, the cancer-masking fashion accomplished 0.90 world accuracy, 0.784 imply intersection over union (mIoU) and imply F1-scores of 0.83 and nil.77 in figuring out nontumor and tumor tissue pixels, respectively. Subsequent, the pipeline used to be run on 2,908 areas (~41 ± 11 areas in keeping with case) randomly decided on from the 71 WSIs in our cohort and routinely remoted 345,038 tumor mobile nuclei (~4,860 nuclei in keeping with case). Nuclear morphology and texture have been quantitated by means of a panel of 63 traits. Distribution of traits in each and every case used to be additional summarized by means of 13 order statistics, yielding 819 options in keeping with case (Fig. 2c and Supplementary Desk 7). A uniform manifold approximation and projection (UMAP) plot published clusters of circumstances with an identical end result (Fig. second) suggesting that one of the most options within the panel endure prognostic possible. The use of the leave-one-out method and the 819 options in keeping with case, we cross-validated seven classification fashions for DS prediction. An RF with an accuracy of 0.66 (95% CI 0.55–0.77) and PPV of 0.76 (95% CI 0.63–0.88) (Fig. 2e) carried out the most productive. All over all validation steps, options realized by means of the tip fashions have been ranked according to the have an effect on on predicting the result and the frequency of prevalence of impactful options measured. Impactful options that came about in a minimum of 10% of validation steps have been thought to be height options. The 17 out of 39 height options to expect survival in Fig. 2f originated from the similar 10 out of 63 nuclear traits in Fig. 2c.Fig. 2: Computational pathology pipeline.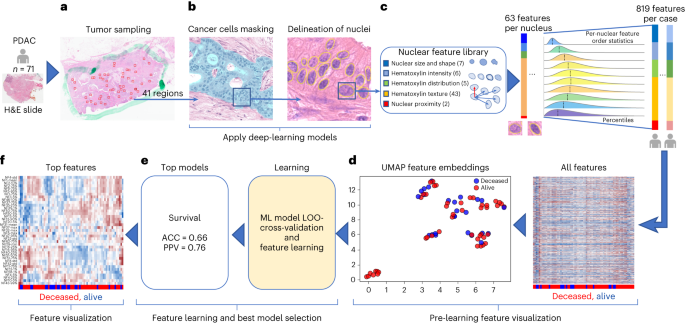 a,b, Pictures of random tumor nests decided on by means of a pathologist in virtual H&E slides (a) are despatched for processing by means of deep-learning fashions to offer a masks of tumor mobile nuclei (b). c, Downstream nuclear function extraction and formation of order statistics of morphology and H&E staining options in nuclei beneath the masks in sufferers from the cohort. d, Affected person-level visualization of extracted options by means of the clustergram (proper) and UMAP function embeddings (left) plots. e, Characteristic studying by means of more than one ML fashions the use of a leave-one-out (LOO) cross-validation technique to determine the fashions that may expect survival with the best possible accuracy. f, Visualization of the tip options realized by means of height survival prediction fashions. The highest options have been decided on according to the function significance realized by means of the fashions.Supply dataTo assess whether or not the computational pathology-based prediction of DS may have the benefit of the inclusion of p.c of stroma or most cancers to stroma ratio in our samples, we carried out our pipeline (Fig. 2b) to the most cancers areas marked by means of our pathologist (W.T.) and measured the share of tumor pixels (pCA), stromal pixels (pST) and the ratio of those two (r = pCA/pST) within the areas (Prolonged Information Fig. 2a,b). No statistically important distinction in pCA (t-test P price = 0.3) and r (t-test P price = 0.257) used to be discovered when tumors related to deficient survival (DS = 1, n = 28) have been in comparison to the ones with higher survival (DS = 0, n = 43). As no distinction used to be noticed, we didn’t incorporate the above options into the computational pathology analyte. Regardless, we discovered that the share of stroma is considerably higher in tissue after neoadjuvant remedy, which will happen following neoadjuvant remedy. Moreover, the share of most cancers used to be smaller in tissue after neoadjuvant remedy, which is the intent of neoadjuvant remedy (Supplementary Desk 8).Multi-omic research suggests hierarchical complementarityThe 6,363 person options from the single-omic resources have been mixed and analyzed the use of seven unbiased ML fashions cross-validated with a leave-one-patient-out method (entire multi-omic function dataset: Desk 1 and Supply Information Fig. 1). Every single-omic supply and multi-omic mixtures have been evaluated the use of all ML fashions. The hyperparameters of each and every fashion have been constant on the preliminary design of the learn about to stop over-optimization and overfitting because of the small cohort measurement. The highest fashion for prediction of DS used to be the multi-omic fashion, which had an accuracy of 0.85 (95% CI 0.73–0.96) and PPV of 0.87 (95% CI 0.75–0.99), adopted by means of single-omic analyte fashions that realized plasma protein, RNA fusions, tissue protein, plasma lipids, medical and surgical pathology, RNA gene expression, computational pathology, DNA CNV, DNA INDELS and DNA SNV options in lowering order of fashion prediction accuracy (Desk 1 and Prolonged Information Fig. 1).The accuracy and PPV efficiency yielded by means of single-omic fashions counsel that each and every single-omic analyte in isolation carries some predictive energy and thus possible medical software. The most productive predictors of DS have been plasma proteins resulting in construction of a fashion with an accuracy of 0.75 (95% CI 0.63–0.86) and PPV of 0.80 (95% CI 0.68–0.92). The fashion studying handiest presurgery CA 19-9 accomplished an accuracy of 0.59 (95% CI 0.47–0.71) and PPV of 0.53 (95% CI 0.40–0.65) and it used to be thought to be the worst amongst all of the single-omic fashions. As noticed within the height two rows of the fashion efficiency (Desk 1), the tip multi-omic fashions outperformed the single-omic ones in accuracy (by means of 10–21%) and PPV (by means of 7–19%) in predicting DS, suggesting complementarity and knowledge achieve throughout analytes when mixed beneath the multi-omic analytical method. Alternatively, the multi-omic fashions had a bigger dispersion of accuracy and PPV, when in comparison to the single-omic fashions (Desk 1 and Prolonged Information Fig. 1) most likely because of the involvement of a far higher set of options to be had for coaching.The 1,024 person analyte mixtures (unmarried and more than one) with all seven modeling methods in keeping with analyte aggregate ended in 7,168 grid seek runs (Fig. 1). To ascertain per-analyte significance, the drop-column significance technique used to be applied and tailored, the place each and every analyte’s set of options have been dropped of their entirety. The use of effects from the 7,168 runs, we evaluated the fashion’s predictive efficiency, analyte composition and have contributions (Fig. 3). Fashions skilled with options from any 2–4 or 9–10 analytes have been inferior in accuracy and PPV to the fashions skilled with options from any 4–8 analytes (Fig. 3a).Fig. 3: Multi-omic efficiency by means of selection of analytes and contribution.
a,b, Pictures of random tumor nests decided on by means of a pathologist in virtual H&E slides (a) are despatched for processing by means of deep-learning fashions to offer a masks of tumor mobile nuclei (b). c, Downstream nuclear function extraction and formation of order statistics of morphology and H&E staining options in nuclei beneath the masks in sufferers from the cohort. d, Affected person-level visualization of extracted options by means of the clustergram (proper) and UMAP function embeddings (left) plots. e, Characteristic studying by means of more than one ML fashions the use of a leave-one-out (LOO) cross-validation technique to determine the fashions that may expect survival with the best possible accuracy. f, Visualization of the tip options realized by means of height survival prediction fashions. The highest options have been decided on according to the function significance realized by means of the fashions.Supply dataTo assess whether or not the computational pathology-based prediction of DS may have the benefit of the inclusion of p.c of stroma or most cancers to stroma ratio in our samples, we carried out our pipeline (Fig. 2b) to the most cancers areas marked by means of our pathologist (W.T.) and measured the share of tumor pixels (pCA), stromal pixels (pST) and the ratio of those two (r = pCA/pST) within the areas (Prolonged Information Fig. 2a,b). No statistically important distinction in pCA (t-test P price = 0.3) and r (t-test P price = 0.257) used to be discovered when tumors related to deficient survival (DS = 1, n = 28) have been in comparison to the ones with higher survival (DS = 0, n = 43). As no distinction used to be noticed, we didn’t incorporate the above options into the computational pathology analyte. Regardless, we discovered that the share of stroma is considerably higher in tissue after neoadjuvant remedy, which will happen following neoadjuvant remedy. Moreover, the share of most cancers used to be smaller in tissue after neoadjuvant remedy, which is the intent of neoadjuvant remedy (Supplementary Desk 8).Multi-omic research suggests hierarchical complementarityThe 6,363 person options from the single-omic resources have been mixed and analyzed the use of seven unbiased ML fashions cross-validated with a leave-one-patient-out method (entire multi-omic function dataset: Desk 1 and Supply Information Fig. 1). Every single-omic supply and multi-omic mixtures have been evaluated the use of all ML fashions. The hyperparameters of each and every fashion have been constant on the preliminary design of the learn about to stop over-optimization and overfitting because of the small cohort measurement. The highest fashion for prediction of DS used to be the multi-omic fashion, which had an accuracy of 0.85 (95% CI 0.73–0.96) and PPV of 0.87 (95% CI 0.75–0.99), adopted by means of single-omic analyte fashions that realized plasma protein, RNA fusions, tissue protein, plasma lipids, medical and surgical pathology, RNA gene expression, computational pathology, DNA CNV, DNA INDELS and DNA SNV options in lowering order of fashion prediction accuracy (Desk 1 and Prolonged Information Fig. 1).The accuracy and PPV efficiency yielded by means of single-omic fashions counsel that each and every single-omic analyte in isolation carries some predictive energy and thus possible medical software. The most productive predictors of DS have been plasma proteins resulting in construction of a fashion with an accuracy of 0.75 (95% CI 0.63–0.86) and PPV of 0.80 (95% CI 0.68–0.92). The fashion studying handiest presurgery CA 19-9 accomplished an accuracy of 0.59 (95% CI 0.47–0.71) and PPV of 0.53 (95% CI 0.40–0.65) and it used to be thought to be the worst amongst all of the single-omic fashions. As noticed within the height two rows of the fashion efficiency (Desk 1), the tip multi-omic fashions outperformed the single-omic ones in accuracy (by means of 10–21%) and PPV (by means of 7–19%) in predicting DS, suggesting complementarity and knowledge achieve throughout analytes when mixed beneath the multi-omic analytical method. Alternatively, the multi-omic fashions had a bigger dispersion of accuracy and PPV, when in comparison to the single-omic fashions (Desk 1 and Prolonged Information Fig. 1) most likely because of the involvement of a far higher set of options to be had for coaching.The 1,024 person analyte mixtures (unmarried and more than one) with all seven modeling methods in keeping with analyte aggregate ended in 7,168 grid seek runs (Fig. 1). To ascertain per-analyte significance, the drop-column significance technique used to be applied and tailored, the place each and every analyte’s set of options have been dropped of their entirety. The use of effects from the 7,168 runs, we evaluated the fashion’s predictive efficiency, analyte composition and have contributions (Fig. 3). Fashions skilled with options from any 2–4 or 9–10 analytes have been inferior in accuracy and PPV to the fashions skilled with options from any 4–8 analytes (Fig. 3a).Fig. 3: Multi-omic efficiency by means of selection of analytes and contribution.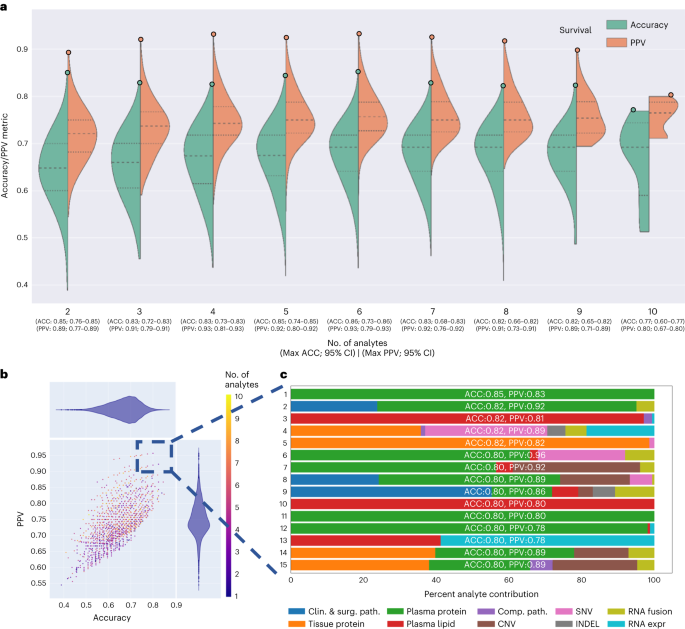 a, Uneven violin plots appearing ACC and PPV distributions for multi-omic survival fashions, segmented by means of selection of analytes within the multi-omic mixtures. Inexperienced and orange dots constitute ACC and PPV of multi-omic analyte mixtures with expanding selection of analytes proper to left at the x axis. b, Multi-omic grid seek fashion effects for DS; selection of analytes 1–10 constitute plasma protein, RNA fusions, tissue protein, lipids, medical and surgical pathology, RNA gene expression, computational pathology, DNA CNV, DNA INDEL and DNA SNV. The y axis presentations PPV and the x axis presentations ACC. c, Best 15 multi-omic fashions for prediction of survival with p.c contribution of each and every person analyte indexed so as of descending accuracy.Supply dataAdditionally, with the drop-column significance method, we have been additionally ready to quantify the significance of each and every analyte class (Supplementary Desk 9) and confirmed that the exclusion of anyone analyte from the learn about most often lowered however didn’t considerably adjust the efficiency; the place the accuracy and PPV for DS prediction have been within the vary of 0.85–0.83 and nil.84–0.83, respectively.Subsequent, we targeted at the height 15 multi-omic fashions for DS prediction (Fig. 3b), that have been the ones with an accuracy >0.80 and PPV > 0.78. We plotted proportions of analyte’s options realized by means of each and every fashion (Fig. 3c) and noticed that the tip fashions had just about an identical accuracies and PPVs, on the other hand the proportions of contributing options various around the height 15 fashions. The primary function contribution used to be from the plasma protein analyte (inexperienced bar, Fig. 3c).Multi-omic fashions supply organic insights into PDACGiven the relative paucity of predictive biomarkers and healing advances in PDAC in comparison to different cancers, a notable exploratory function of our learn about used to be to evaluate whether or not our platform can determine possible pathways and objectives of remedy. The use of a differentially expressed function set, we have been ready to establish options to check function Spearman correlations and the significance for all analyte options (Fig. 4a). By means of comparing analyte contribution for each and every fashion, it used to be conceivable to generate ontology visualizations for protein, DNA and RNA as proven for the tip multi-omic fashions for DS (Fig. 4b).Fig. 4: Organic relevance of height options in muti-omic fashion and clustering.
a, Uneven violin plots appearing ACC and PPV distributions for multi-omic survival fashions, segmented by means of selection of analytes within the multi-omic mixtures. Inexperienced and orange dots constitute ACC and PPV of multi-omic analyte mixtures with expanding selection of analytes proper to left at the x axis. b, Multi-omic grid seek fashion effects for DS; selection of analytes 1–10 constitute plasma protein, RNA fusions, tissue protein, lipids, medical and surgical pathology, RNA gene expression, computational pathology, DNA CNV, DNA INDEL and DNA SNV. The y axis presentations PPV and the x axis presentations ACC. c, Best 15 multi-omic fashions for prediction of survival with p.c contribution of each and every person analyte indexed so as of descending accuracy.Supply dataAdditionally, with the drop-column significance method, we have been additionally ready to quantify the significance of each and every analyte class (Supplementary Desk 9) and confirmed that the exclusion of anyone analyte from the learn about most often lowered however didn’t considerably adjust the efficiency; the place the accuracy and PPV for DS prediction have been within the vary of 0.85–0.83 and nil.84–0.83, respectively.Subsequent, we targeted at the height 15 multi-omic fashions for DS prediction (Fig. 3b), that have been the ones with an accuracy >0.80 and PPV > 0.78. We plotted proportions of analyte’s options realized by means of each and every fashion (Fig. 3c) and noticed that the tip fashions had just about an identical accuracies and PPVs, on the other hand the proportions of contributing options various around the height 15 fashions. The primary function contribution used to be from the plasma protein analyte (inexperienced bar, Fig. 3c).Multi-omic fashions supply organic insights into PDACGiven the relative paucity of predictive biomarkers and healing advances in PDAC in comparison to different cancers, a notable exploratory function of our learn about used to be to evaluate whether or not our platform can determine possible pathways and objectives of remedy. The use of a differentially expressed function set, we have been ready to establish options to check function Spearman correlations and the significance for all analyte options (Fig. 4a). By means of comparing analyte contribution for each and every fashion, it used to be conceivable to generate ontology visualizations for protein, DNA and RNA as proven for the tip multi-omic fashions for DS (Fig. 4b).Fig. 4: Organic relevance of height options in muti-omic fashion and clustering.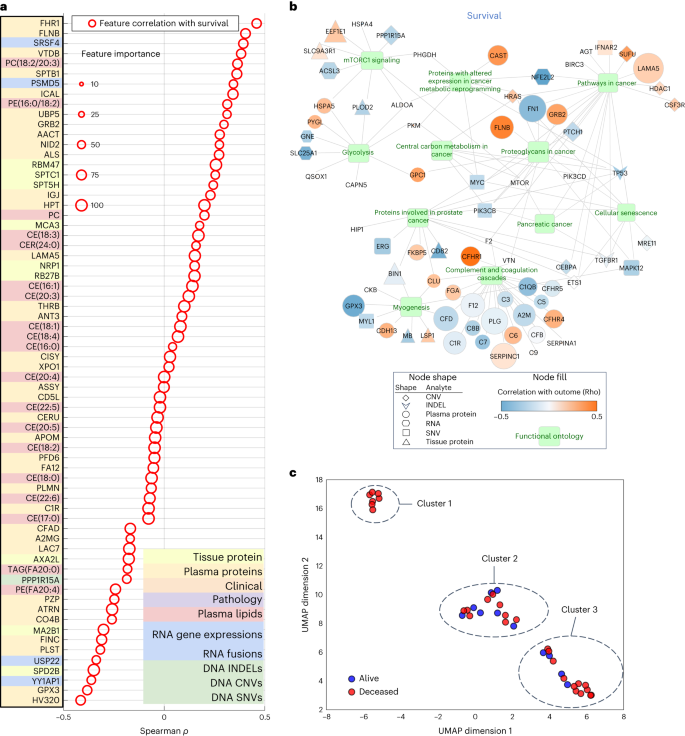 a, Spearman correlation of height multi-omic options with DS. Measurement represents a function’s relative significance to the tip multi-omic fashion; the crimson colour signifies whether or not the function significance relates to DS. b, Gene Ontology community visualization for many informative options from the multi-omic fashions. Decided on practical pathways containing gene units from multi-omic analytes are displayed as inexperienced nodes, with related genes and measured analyte sorts represented by means of a particular form (according to analyte) and coloured in line with the energy of a given analyte’s correlation to the result variable of DS. Measurement of a given analyte node is relative to the frequency with which that analyte used to be decided on for the fashions, with higher analytes extra constantly decided on and no visual node indicating that the analyte used to be now not decided on as essential for the DS end result displayed. c, UMAP clusters of sufferers the use of molecular signatures consisting of all 6,363 multi-omic options, coloured by means of survival.Supply datamTOR signaling, a recognized pathway in lots of tumors34,35 together with PDAC, used to be discovered within the Gene Ontology community visualizations of the tip multi-omic models36 (Fig. 4b). It’s been focused in PDAC by myself and together with different agents37 with combined effects. Aside from mTOR, our Gene Ontology community visualizations published different clinically and biologically related pathways in PDAC, together with glycolysis and cell metabolism38,39.To inspect the connection of tumor to end result heterogeneity, all 6,363 options throughout all analytes have been used to create patient-level clustering according to multi-omic molecular signatures and categorised for DS (Fig. 4c). Cluster 1 represents sufferers homogeneous for his or her medical end result (all deceased). To higher perceive the affiliation of the heterogenous clusters, (2 and three), with different medical and computational pathology options, we in comparison the expression of a function in a single cluster to that within the two different clusters mixed the use of t-test or Fisher’s examine. This research published proportions of related options (P < 0.05) in each and every analyte (Supplementary Desk 10), the place with the exception of for computational pathology, no different analyte contained options that have been found in all 3 pair-wise comparisons. Therefore, we used one-way research of variance, which recognized 8 differentially expressed computational pathology options (Supplementary Desk 11). Those 8 options have been then analyzed by means of the Tukey–Kramer examine for more than one comparisons. No function used to be considerably other between the 3 clusters, however there have been options that differed between two clusters. Moreover, hierarchical clustering of 39 topics characterised by means of the 8 computational pathology options (Prolonged Information Fig. 3) prompt that they strongly contributed to the formation of clusters 1, 2 and three. In combination, those findings counsel that with extra sufferers and with potential iterative research over the years, our method will lead to regularly extra correct predictions particularly for sufferers who have compatibility club in particular clusters (as an example, cluster 1) and deeper perception into what options are crucial to person affected person clusters.The parsimonious multi-omic fashions for illness survivalThe complementarity of analytes noticed in multi-omic fashions in Desk 1 and Fig. 3, prompt {that a} parsimonious multi-omic fashion providing an identical predictive efficiency to fashions with higher and extra complicated analyte compositions may well be advanced. If true, the worldwide public well being and societal have an effect on could be consequential as it might doubtlessly start the method of democratizing precision most cancers drugs, particularly to spaces of the arena with restricted monetary and technical healthcare sources. To check this speculation, we began with all the multi-omic function house of 6,363 options and skilled an RF fashion for DS using a recursive function removing (RFE) technique such that at each and every step the least-informative options have been eradicated from additional fashion iterations (Fig. 5a). Maximum particularly, Fig. 5a highlights the inflection level of the ‘parsimonious fashion’ location at the curve (accuracy of 0.85, PPV of 0.85) studying handiest 589 multi-omic options. Additional, the contribution of respective analytes to the parsimonious fashion stays most commonly solid throughout iterations after the inflection level, with plasma lipids and RNA being probably the most related; on the other hand, be aware that plasma (proteins or lipids) by myself can give correct prediction with fewer options. This opens the likelihood {that a} screening of plasma may ultimately be used for decision-making relating to pancreatic surgical procedure.Fig. 5: Efficiency of parsimonious multi-omic fashions and analyte contribution for illness survival.
a, Spearman correlation of height multi-omic options with DS. Measurement represents a function’s relative significance to the tip multi-omic fashion; the crimson colour signifies whether or not the function significance relates to DS. b, Gene Ontology community visualization for many informative options from the multi-omic fashions. Decided on practical pathways containing gene units from multi-omic analytes are displayed as inexperienced nodes, with related genes and measured analyte sorts represented by means of a particular form (according to analyte) and coloured in line with the energy of a given analyte’s correlation to the result variable of DS. Measurement of a given analyte node is relative to the frequency with which that analyte used to be decided on for the fashions, with higher analytes extra constantly decided on and no visual node indicating that the analyte used to be now not decided on as essential for the DS end result displayed. c, UMAP clusters of sufferers the use of molecular signatures consisting of all 6,363 multi-omic options, coloured by means of survival.Supply datamTOR signaling, a recognized pathway in lots of tumors34,35 together with PDAC, used to be discovered within the Gene Ontology community visualizations of the tip multi-omic models36 (Fig. 4b). It’s been focused in PDAC by myself and together with different agents37 with combined effects. Aside from mTOR, our Gene Ontology community visualizations published different clinically and biologically related pathways in PDAC, together with glycolysis and cell metabolism38,39.To inspect the connection of tumor to end result heterogeneity, all 6,363 options throughout all analytes have been used to create patient-level clustering according to multi-omic molecular signatures and categorised for DS (Fig. 4c). Cluster 1 represents sufferers homogeneous for his or her medical end result (all deceased). To higher perceive the affiliation of the heterogenous clusters, (2 and three), with different medical and computational pathology options, we in comparison the expression of a function in a single cluster to that within the two different clusters mixed the use of t-test or Fisher’s examine. This research published proportions of related options (P < 0.05) in each and every analyte (Supplementary Desk 10), the place with the exception of for computational pathology, no different analyte contained options that have been found in all 3 pair-wise comparisons. Therefore, we used one-way research of variance, which recognized 8 differentially expressed computational pathology options (Supplementary Desk 11). Those 8 options have been then analyzed by means of the Tukey–Kramer examine for more than one comparisons. No function used to be considerably other between the 3 clusters, however there have been options that differed between two clusters. Moreover, hierarchical clustering of 39 topics characterised by means of the 8 computational pathology options (Prolonged Information Fig. 3) prompt that they strongly contributed to the formation of clusters 1, 2 and three. In combination, those findings counsel that with extra sufferers and with potential iterative research over the years, our method will lead to regularly extra correct predictions particularly for sufferers who have compatibility club in particular clusters (as an example, cluster 1) and deeper perception into what options are crucial to person affected person clusters.The parsimonious multi-omic fashions for illness survivalThe complementarity of analytes noticed in multi-omic fashions in Desk 1 and Fig. 3, prompt {that a} parsimonious multi-omic fashion providing an identical predictive efficiency to fashions with higher and extra complicated analyte compositions may well be advanced. If true, the worldwide public well being and societal have an effect on could be consequential as it might doubtlessly start the method of democratizing precision most cancers drugs, particularly to spaces of the arena with restricted monetary and technical healthcare sources. To check this speculation, we began with all the multi-omic function house of 6,363 options and skilled an RF fashion for DS using a recursive function removing (RFE) technique such that at each and every step the least-informative options have been eradicated from additional fashion iterations (Fig. 5a). Maximum particularly, Fig. 5a highlights the inflection level of the ‘parsimonious fashion’ location at the curve (accuracy of 0.85, PPV of 0.85) studying handiest 589 multi-omic options. Additional, the contribution of respective analytes to the parsimonious fashion stays most commonly solid throughout iterations after the inflection level, with plasma lipids and RNA being probably the most related; on the other hand, be aware that plasma (proteins or lipids) by myself can give correct prediction with fewer options. This opens the likelihood {that a} screening of plasma may ultimately be used for decision-making relating to pancreatic surgical procedure.Fig. 5: Efficiency of parsimonious multi-omic fashions and analyte contribution for illness survival.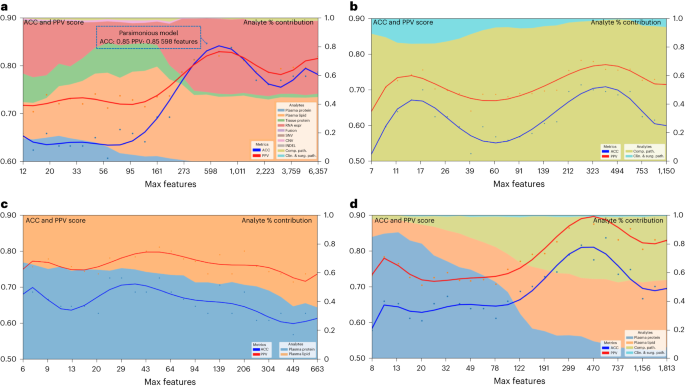 a, Parsimonious fashion of all multi-omic options and entire dataset. The blue dotted line field signifies the parsimonious fashion on the inflection level. b, Scientific and surgical pathology and computational pathology analytes handiest. c, All plasma analytes (lipidomics and protein) handiest. d, All medical and surgical pathology, computational pathology and plasma analytes (lipidomics and protein) handiest. Left y axis presentations accuracy and PPV rating: multi-omic fashion efficiency throughout function aid steps by means of limiting the utmost selectable options throughout fashion coaching. The x axis presentations the selection of most options at each and every aid step. The best y axis presentations the analyte p.c (%) contribution: each and every analyte’s aggregated absolute function weight contribution at each and every function aid step.Supply dataTrying to inspect the possibility of this method for eventual globalization of precision oncology, we assessed particular restricted analyte mixtures and have units that may be carried out to our parsimonious fashion. Those analytes have been decided on according to standards of usual availability (pathology specimens or medical information together with surgical pathology) or simply acquired (plasma lipids or proteins) as a part of the diagnostic workup. The use of this method, we recognized correct parsimonious fashions that realized options from medical, surgical pathology and computational pathology analytes (Fig. 5b), all plasma analytes (lipidomics and protein) (Fig. 5c) and medical, mixed with computational pathology and plasma analytes (Fig. 5d) and which had an identical accuracy and PPV to the fashions that realized options from all of the set of 6,363 options in Fig. 5a.Validation of RNA markers as predictors of survivalWhole-transcriptome sequencing and research used to be carried out on 57 samples from our pilot cohort (Supplementary Desk 4). Using L1-normalized RF modeling, RNA gene transcripts considerably (P ≤ 0.05) predicting survival (n = 79) have been used to expand gene signatures for stepped forward (sure Pearson and Spearman rho for survival, n = 40 genes) and for deficient (adverse Pearson and Spearman rho for survival, n = 39 genes) survival (Supplementary Desk 12). Those two signatures have been evaluated in an unbiased dataset of 177 PDAC patients40 for his or her skill to stratify DS. Top rating of the signature composed of genes whose expression used to be related to deficient diagnosis in our information (n = 39) used to be additionally related to deficient DS on this set (danger ratio (HR) = 2.17, (1.28–3.66), log-rank P = 0.0031) (Prolonged Information Fig. 4a), while that of genes whose expression used to be outlined as a excellent prognostic in our information (n = 40), had a pattern towards stepped forward DS (HR = 0.74 (0.49–1.12), log-rank P = 0.15) (Prolonged Information Fig. 4b). We additionally carried out gene set enrichment research at the RNA transcripts used within the two signatures above (n = 79). Enrichr41 discovered a lot of important pathways (Supplementary Desk 13) implicated in PDAC resistance and treatment-targeting, together with interferon signaling, AMP-activated protein kinase (AMPK) and CXCR4 signaling pathways42,43,44,45. In combination, those information independently validate the medical relevance of our RNA expression discoveries.Validation of multi-omic fashions as predictors of survivalTo additional validate our single-omic, multi-omic and parsimonious analytes for DS prediction, we evaluated their predictive efficiency at the TCGA dataset, containing 157 evaluable samples that had a minimum of one analyte sort (Supplementary Desk 1)46. As TCGA has information handiest on DNA, RNA, virtual H&E slides and medical analytes, our modeling had a discounted set of three,423 overall options in comparison to the 6,363 in our MT-Pilot cohort (Desk 1 and Fig. 1e). The entire 3,423 analyte fashion had an accuracy and PPV of 0.94 (95 CI 0.83–1.00) and nil.95 (95% CI 0.84–1.00) (Desk 2 and Supplementary Desk 14) for DS prediction with computational pathology, DNA SNVs and RNA gene expressions acting strongly in single-omic validation of DS (Desk 2 and Supplementary Desk 14).Desk 2 Best single-omic and multi-omic efficiency for predicting illness survival in PDAC: learn about validation cohortsNext, we tested the validity of our multi-omic parsimonious fashion at the TCGA dataset. As a result of this cohort had an general lowered analyte set, we used an RFE technique to retrain a RF fashion for DS on our cohort (MT-Pilot) and made up our minds that the optimum (height of top) parsimonious fashion hired 202 options out of three,423 and had accuracy and PPV of 0.74 (0.63–0.85) and nil.77 (0.65–0.89), respectively (Prolonged Information Fig. 4c). Significantly, when the fashion used to be carried out to those identical 202 options (Supplementary Desk 15) within the TCGA dataset, it yielded an accuracy of 0.88 and PPV of 0.95 for DS prediction. Moreover, in each our MT-Pilot cohort and the TCGA cohort, computational pathology and RNA gene expression have been discovered to be number one analytes realized by means of the DS predicting fashions, with CNV and the medical analyte offering minor further development (Prolonged Information Fig. 4c). The sign dominance of RNA isn’t pushed by means of expression of any unmarried gene, however by means of a particular set of genes.As TCGA lacked tissue proteomic point information, we sought an exterior dataset with tissue protein information, together with different crucial single-omic informative analytes similar to DNA, RNA and medical information. We discovered an unbiased publicly to be had dataset14 named JHU cohort 1 that met those standards. With DNA, RNA, medical information and tissue protein analytes from our MT-Pilot cohort serving as the educational set, we skilled an L1-normalized RF fashion and carried out it to this validation examine set. This fashion predicted DS with an accuracy and PPV of 0.89 (95% CI 0.83–0.95) and nil.91 (95% CI 0.85–0.98), respectively (Desk 2 and Supplementary Desk 14). Whilst a fashion skilled at the tissue protein as a single-omic analyte had an accuracy and PPV of 0.56 (95% CI 0.50–0.63) and nil.53 (95% CI 0.47–0.60) within the JHU cohort 1 (Desk 2 and Supplementary Desk 14), addition of DNA, RNA and medical analytes stepped forward the predictive efficiency of the fashion and validated the multi-omics method.Validation of plasma proteins as a preoperative biomarkerThrough our multi-omic and parsimonious modeling of the MT-Pilot cohort, we found out that plasma protein is an analyte that gives now not handiest correct prediction of DS in PDAC, however does so with the fewest options in comparison to different analytes. On account of those findings, in addition to the deficient efficiency of CA 19-9 as a preoperative marker for decision-making relating to the advantage of surgical procedure, we subsequent sought to validate our findings only on analytes that will be to be had to the medical practitioner prior to surgical procedure.But even so the TCGA and JHU cohort 1, we applied two extra cohorts; JHU cohort 2 and the MGH cohort (Supplementary Desk 1). They incorporated an identical degree I/II resected PDAC with medical and demographic information gathered longitudinally and preoperative plasma samples, together with CA 19-9 acquired and analyzed as described above. Utility of the L1-normalized RF fashion skilled at the MT-Pilot information at the two cohorts confirmed that plasma proteins remained extremely predictive of DS in each validation cohorts, with accuracy and PPV of 0.98 (95% CI 0.83–1.00) 0.92 (95% CI 0.79–1.00), respectively in JHU cohort 2 and nil.89 (95% CI 0.76–1.00) 0.80 (95% CI 0.69–0.91), respectively within the MGH cohort (Desk 2 and Supplementary Desk 14). The addition of medical information to plasma protein improves the multi-omics fashion for DS prediction. Total, preoperative plasma protein used to be extremely predictive of DS amongst 3 separate unbiased datasets and equipped a novel preoperative biomarker with much better predictive efficiency than mechanically applied CA 19-9 (Desk 2 and Supplementary Desk 14).
a, Parsimonious fashion of all multi-omic options and entire dataset. The blue dotted line field signifies the parsimonious fashion on the inflection level. b, Scientific and surgical pathology and computational pathology analytes handiest. c, All plasma analytes (lipidomics and protein) handiest. d, All medical and surgical pathology, computational pathology and plasma analytes (lipidomics and protein) handiest. Left y axis presentations accuracy and PPV rating: multi-omic fashion efficiency throughout function aid steps by means of limiting the utmost selectable options throughout fashion coaching. The x axis presentations the selection of most options at each and every aid step. The best y axis presentations the analyte p.c (%) contribution: each and every analyte’s aggregated absolute function weight contribution at each and every function aid step.Supply dataTrying to inspect the possibility of this method for eventual globalization of precision oncology, we assessed particular restricted analyte mixtures and have units that may be carried out to our parsimonious fashion. Those analytes have been decided on according to standards of usual availability (pathology specimens or medical information together with surgical pathology) or simply acquired (plasma lipids or proteins) as a part of the diagnostic workup. The use of this method, we recognized correct parsimonious fashions that realized options from medical, surgical pathology and computational pathology analytes (Fig. 5b), all plasma analytes (lipidomics and protein) (Fig. 5c) and medical, mixed with computational pathology and plasma analytes (Fig. 5d) and which had an identical accuracy and PPV to the fashions that realized options from all of the set of 6,363 options in Fig. 5a.Validation of RNA markers as predictors of survivalWhole-transcriptome sequencing and research used to be carried out on 57 samples from our pilot cohort (Supplementary Desk 4). Using L1-normalized RF modeling, RNA gene transcripts considerably (P ≤ 0.05) predicting survival (n = 79) have been used to expand gene signatures for stepped forward (sure Pearson and Spearman rho for survival, n = 40 genes) and for deficient (adverse Pearson and Spearman rho for survival, n = 39 genes) survival (Supplementary Desk 12). Those two signatures have been evaluated in an unbiased dataset of 177 PDAC patients40 for his or her skill to stratify DS. Top rating of the signature composed of genes whose expression used to be related to deficient diagnosis in our information (n = 39) used to be additionally related to deficient DS on this set (danger ratio (HR) = 2.17, (1.28–3.66), log-rank P = 0.0031) (Prolonged Information Fig. 4a), while that of genes whose expression used to be outlined as a excellent prognostic in our information (n = 40), had a pattern towards stepped forward DS (HR = 0.74 (0.49–1.12), log-rank P = 0.15) (Prolonged Information Fig. 4b). We additionally carried out gene set enrichment research at the RNA transcripts used within the two signatures above (n = 79). Enrichr41 discovered a lot of important pathways (Supplementary Desk 13) implicated in PDAC resistance and treatment-targeting, together with interferon signaling, AMP-activated protein kinase (AMPK) and CXCR4 signaling pathways42,43,44,45. In combination, those information independently validate the medical relevance of our RNA expression discoveries.Validation of multi-omic fashions as predictors of survivalTo additional validate our single-omic, multi-omic and parsimonious analytes for DS prediction, we evaluated their predictive efficiency at the TCGA dataset, containing 157 evaluable samples that had a minimum of one analyte sort (Supplementary Desk 1)46. As TCGA has information handiest on DNA, RNA, virtual H&E slides and medical analytes, our modeling had a discounted set of three,423 overall options in comparison to the 6,363 in our MT-Pilot cohort (Desk 1 and Fig. 1e). The entire 3,423 analyte fashion had an accuracy and PPV of 0.94 (95 CI 0.83–1.00) and nil.95 (95% CI 0.84–1.00) (Desk 2 and Supplementary Desk 14) for DS prediction with computational pathology, DNA SNVs and RNA gene expressions acting strongly in single-omic validation of DS (Desk 2 and Supplementary Desk 14).Desk 2 Best single-omic and multi-omic efficiency for predicting illness survival in PDAC: learn about validation cohortsNext, we tested the validity of our multi-omic parsimonious fashion at the TCGA dataset. As a result of this cohort had an general lowered analyte set, we used an RFE technique to retrain a RF fashion for DS on our cohort (MT-Pilot) and made up our minds that the optimum (height of top) parsimonious fashion hired 202 options out of three,423 and had accuracy and PPV of 0.74 (0.63–0.85) and nil.77 (0.65–0.89), respectively (Prolonged Information Fig. 4c). Significantly, when the fashion used to be carried out to those identical 202 options (Supplementary Desk 15) within the TCGA dataset, it yielded an accuracy of 0.88 and PPV of 0.95 for DS prediction. Moreover, in each our MT-Pilot cohort and the TCGA cohort, computational pathology and RNA gene expression have been discovered to be number one analytes realized by means of the DS predicting fashions, with CNV and the medical analyte offering minor further development (Prolonged Information Fig. 4c). The sign dominance of RNA isn’t pushed by means of expression of any unmarried gene, however by means of a particular set of genes.As TCGA lacked tissue proteomic point information, we sought an exterior dataset with tissue protein information, together with different crucial single-omic informative analytes similar to DNA, RNA and medical information. We discovered an unbiased publicly to be had dataset14 named JHU cohort 1 that met those standards. With DNA, RNA, medical information and tissue protein analytes from our MT-Pilot cohort serving as the educational set, we skilled an L1-normalized RF fashion and carried out it to this validation examine set. This fashion predicted DS with an accuracy and PPV of 0.89 (95% CI 0.83–0.95) and nil.91 (95% CI 0.85–0.98), respectively (Desk 2 and Supplementary Desk 14). Whilst a fashion skilled at the tissue protein as a single-omic analyte had an accuracy and PPV of 0.56 (95% CI 0.50–0.63) and nil.53 (95% CI 0.47–0.60) within the JHU cohort 1 (Desk 2 and Supplementary Desk 14), addition of DNA, RNA and medical analytes stepped forward the predictive efficiency of the fashion and validated the multi-omics method.Validation of plasma proteins as a preoperative biomarkerThrough our multi-omic and parsimonious modeling of the MT-Pilot cohort, we found out that plasma protein is an analyte that gives now not handiest correct prediction of DS in PDAC, however does so with the fewest options in comparison to different analytes. On account of those findings, in addition to the deficient efficiency of CA 19-9 as a preoperative marker for decision-making relating to the advantage of surgical procedure, we subsequent sought to validate our findings only on analytes that will be to be had to the medical practitioner prior to surgical procedure.But even so the TCGA and JHU cohort 1, we applied two extra cohorts; JHU cohort 2 and the MGH cohort (Supplementary Desk 1). They incorporated an identical degree I/II resected PDAC with medical and demographic information gathered longitudinally and preoperative plasma samples, together with CA 19-9 acquired and analyzed as described above. Utility of the L1-normalized RF fashion skilled at the MT-Pilot information at the two cohorts confirmed that plasma proteins remained extremely predictive of DS in each validation cohorts, with accuracy and PPV of 0.98 (95% CI 0.83–1.00) 0.92 (95% CI 0.79–1.00), respectively in JHU cohort 2 and nil.89 (95% CI 0.76–1.00) 0.80 (95% CI 0.69–0.91), respectively within the MGH cohort (Desk 2 and Supplementary Desk 14). The addition of medical information to plasma protein improves the multi-omics fashion for DS prediction. Total, preoperative plasma protein used to be extremely predictive of DS amongst 3 separate unbiased datasets and equipped a novel preoperative biomarker with much better predictive efficiency than mechanically applied CA 19-9 (Desk 2 and Supplementary Desk 14).
The Molecular Dual artificial-intelligence platform integrates multi-omic information to expect results for pancreatic adenocarcinoma sufferers – Nature Most cancers