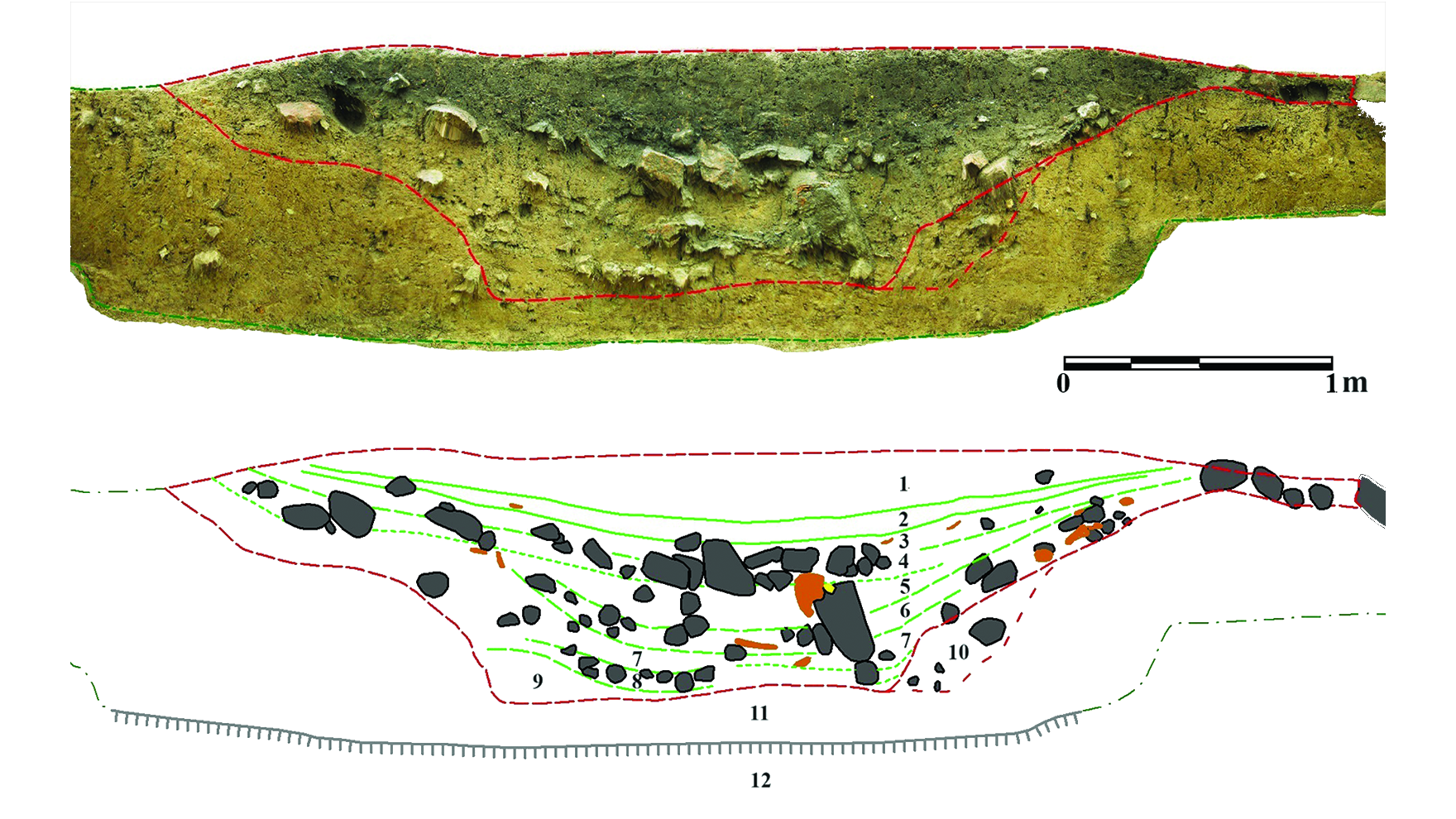
Creating an epigenetic screening platform in human T cellsStaphylococcus aureus Cas9 (SaCas9) has been widely used for genome enhancing in vivo as its compact measurement (3,159 bp) relative to the normal Streptococcus pyogenes Cas9 (SpCas9) permits packaging into adeno-associated virus26,27,28. Then again, SaCas9 has now not been extensively used for focused gene regulation29,30 or within the context of an epigenome enhancing display screen. To facilitate supply to human T cells, we conscientiously characterised the task of dSaCas9 as a repressor or activator the usage of a number of promoter tiling information RNA (gRNA) monitors in each number one human T cells and the Jurkat mobile line (Prolonged Information Figs. 1–3, Supplementary Fig. 1 and Supplementary Word 1). Jointly, this paintings demonstrated that dSaCas9 can potently silence or turn on goal gene expression and knowledgeable gRNA design laws.CRISPRi/a monitors establish regulators of human T mobile stateWe sought to interrogate the results of repressing or activating genes encoding TFs and epigenetic modifiers on T mobile state. We designed a gRNA library concentrating on 120 TFs and epigenetic modifiers related to T mobile state (Supplementary Fig. 3 and Supplementary Word 2). To stumble on whether or not particular gene perturbations altered T mobile state, we used CCR7 expression as our display screen readout (Fig. 1a and Supplementary Fig. 4). CCR7 is a well-characterized T mobile marker and is very expressed in particular T mobile subsets similar to naive, stem-cell reminiscence and central reminiscence T cells31. We hypothesized it might permit us to seize extra refined adjustments in T mobile state than phenotypic readouts similar to proliferation or cytokine manufacturing.Fig. 1: CRISPR interference or activation genetic monitors establish transcriptional and epigenetic regulators of human CD8+ T mobile state.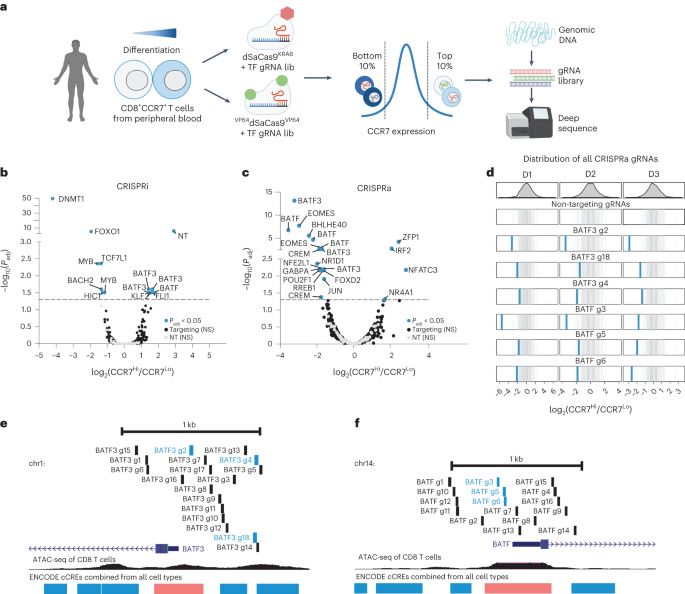 a, Schematic of CRISPRi/a monitors with TF gRNA library (lib). b,c, Importance (Padj) as opposed to fold exchange in gRNA abundance between CCR7HIGH and CCR7LOW populations for CRISPRi (b) and CRISPRa (c) monitors. gRNA enrichment used to be outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. d, Fold exchange of BATF3 and BATF CRISPRa gRNA hits for each and every donor (D1-D3). Blue traces constitute BATF3 or BATF gRNAs and grey traces constitute the distribution of 120 non-targeting (NT) keep watch over gRNAs. e,f, All BATF3 (e) and BATF (f) CRISPRa gRNAs in gRNA library relative to TSS, chromatin accessibility and ENCODE candidate cis-regulatory parts (cCREs). Blue and black traces constitute gRNA hits and nonsignificant gRNAs, respectively. cCRE tracks are overlaid for visualisation of promoter-like parts (pink) and enhancer-like parts (blue).The CRISPRi display screen recovered many canonical regulators of reminiscence T cells together with FOXO1 (ref. 32), MYB33 and BACH2 (ref. 34)—all of which when silenced resulted in lowered expression of CCR7, indicative of T mobile differentiation in opposition to effector T cells (Fig. 1b and Supplementary Fig. 5a). Apparently, essentially the most vital hit from the CRISPRi display screen used to be the gene encoding the upkeep DNA methyltransferase DNMT1. Genetic disruption of each TET2 and DNMT3A, which encode for proteins that control DNA methylation in reverse instructions, can make stronger the healing attainable of T cells19,20. There used to be a unmarried nontargeting (NT) gRNA (1/120) hit within the CRISPRi display screen. The similar NT gRNA emerged as successful in more than one monitors the usage of CCR7 because the readout, suggesting an actual off-target impact.The CRISPRa display screen additionally recognized a number of TFs which have been implicated in CD8+ T mobile differentiation and serve as similar to EOMES35, BATF13 and JUN12 (Fig. 1c). Importantly, gRNA enrichment used to be constant around the 3 donors and now not a operate of the choice of gRNAs concentrating on each and every gene (Fig. 1d and Supplementary Fig. 5). A couple of gRNAs concentrating on BATF and BATF3 have been enriched in reciprocal instructions throughout CRISPRi and CRISPRa monitors, and BATF and BATF3 have been a few of the best hits in gene-level analyses, highlighting the facility of coupling loss- or gain-of-function perturbations (Supplementary Desk 2). The BATF and BATF3 gRNA hits within the CRISPRa display screen usually colocalized to areas upstream of the promoter and close to the summits of obtainable chromatin (Fig. 1e,f).scRNA-seq characterization of transcriptional regulatorsWe subsequent characterised the transcriptomic results of each and every candidate gene recognized from our CRISPRi or CRISPRa monitors the usage of single-cell RNA sequencing (scRNA-seq). We cloned the union set of gRNA hits throughout CRISPRi/a monitors (32 gRNAs) and eight NT gRNAs into each CRISPRi and CRISPRa plasmids (Supplementary Desk 3). We then adopted the similar workflow because the sort-based monitors, however as an alternative of sorting the cells in keeping with CCR7 expression, we profiled the transcriptomes and gRNA id of ~60,000 cells throughout 3 donors for each and every display screen. We aggregated the cells in keeping with gRNA project and when put next the transcriptional profile of cells with the similar gRNA to nonperturbed cells (Supplementary Fig. 6 and Supplementary Word 3).First, we fascinated with CCR7 expression to validate the effects from our CRISPRi/a monitors (Fig. 2a,b). Kind of part of the gRNA hits affected CCR7 expression, and the rank order used to be very similar to the sort-based monitors. As an example, each assays knowledgeable that focused silencing of DNMT1 or FOXO1 significantly lowered CCR7 expression ranges, which used to be additional showed via particular person gRNA validations (Supplementary Fig. 7a,b). The gRNA hits that didn’t validate within the scRNA-seq characterization have been represented by means of fewer cells than validated gRNAs, reaffirming that upper gRNA protection is helping to unravel extra refined adjustments in gene expression36 (Supplementary Fig. 7c and Supplementary Word 4). Along with confirming gRNA results on CCR7 expression, the actual adverse charges have been prime for each CRISPRi (96%) and CRISPRa (82%), demonstrating the specificity of those sort-based monitors (Fig. 2a,b).Fig. 2: scRNA-seq characterization of candidate genes.
a, Schematic of CRISPRi/a monitors with TF gRNA library (lib). b,c, Importance (Padj) as opposed to fold exchange in gRNA abundance between CCR7HIGH and CCR7LOW populations for CRISPRi (b) and CRISPRa (c) monitors. gRNA enrichment used to be outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. d, Fold exchange of BATF3 and BATF CRISPRa gRNA hits for each and every donor (D1-D3). Blue traces constitute BATF3 or BATF gRNAs and grey traces constitute the distribution of 120 non-targeting (NT) keep watch over gRNAs. e,f, All BATF3 (e) and BATF (f) CRISPRa gRNAs in gRNA library relative to TSS, chromatin accessibility and ENCODE candidate cis-regulatory parts (cCREs). Blue and black traces constitute gRNA hits and nonsignificant gRNAs, respectively. cCRE tracks are overlaid for visualisation of promoter-like parts (pink) and enhancer-like parts (blue).The CRISPRi display screen recovered many canonical regulators of reminiscence T cells together with FOXO1 (ref. 32), MYB33 and BACH2 (ref. 34)—all of which when silenced resulted in lowered expression of CCR7, indicative of T mobile differentiation in opposition to effector T cells (Fig. 1b and Supplementary Fig. 5a). Apparently, essentially the most vital hit from the CRISPRi display screen used to be the gene encoding the upkeep DNA methyltransferase DNMT1. Genetic disruption of each TET2 and DNMT3A, which encode for proteins that control DNA methylation in reverse instructions, can make stronger the healing attainable of T cells19,20. There used to be a unmarried nontargeting (NT) gRNA (1/120) hit within the CRISPRi display screen. The similar NT gRNA emerged as successful in more than one monitors the usage of CCR7 because the readout, suggesting an actual off-target impact.The CRISPRa display screen additionally recognized a number of TFs which have been implicated in CD8+ T mobile differentiation and serve as similar to EOMES35, BATF13 and JUN12 (Fig. 1c). Importantly, gRNA enrichment used to be constant around the 3 donors and now not a operate of the choice of gRNAs concentrating on each and every gene (Fig. 1d and Supplementary Fig. 5). A couple of gRNAs concentrating on BATF and BATF3 have been enriched in reciprocal instructions throughout CRISPRi and CRISPRa monitors, and BATF and BATF3 have been a few of the best hits in gene-level analyses, highlighting the facility of coupling loss- or gain-of-function perturbations (Supplementary Desk 2). The BATF and BATF3 gRNA hits within the CRISPRa display screen usually colocalized to areas upstream of the promoter and close to the summits of obtainable chromatin (Fig. 1e,f).scRNA-seq characterization of transcriptional regulatorsWe subsequent characterised the transcriptomic results of each and every candidate gene recognized from our CRISPRi or CRISPRa monitors the usage of single-cell RNA sequencing (scRNA-seq). We cloned the union set of gRNA hits throughout CRISPRi/a monitors (32 gRNAs) and eight NT gRNAs into each CRISPRi and CRISPRa plasmids (Supplementary Desk 3). We then adopted the similar workflow because the sort-based monitors, however as an alternative of sorting the cells in keeping with CCR7 expression, we profiled the transcriptomes and gRNA id of ~60,000 cells throughout 3 donors for each and every display screen. We aggregated the cells in keeping with gRNA project and when put next the transcriptional profile of cells with the similar gRNA to nonperturbed cells (Supplementary Fig. 6 and Supplementary Word 3).First, we fascinated with CCR7 expression to validate the effects from our CRISPRi/a monitors (Fig. 2a,b). Kind of part of the gRNA hits affected CCR7 expression, and the rank order used to be very similar to the sort-based monitors. As an example, each assays knowledgeable that focused silencing of DNMT1 or FOXO1 significantly lowered CCR7 expression ranges, which used to be additional showed via particular person gRNA validations (Supplementary Fig. 7a,b). The gRNA hits that didn’t validate within the scRNA-seq characterization have been represented by means of fewer cells than validated gRNAs, reaffirming that upper gRNA protection is helping to unravel extra refined adjustments in gene expression36 (Supplementary Fig. 7c and Supplementary Word 4). Along with confirming gRNA results on CCR7 expression, the actual adverse charges have been prime for each CRISPRi (96%) and CRISPRa (82%), demonstrating the specificity of those sort-based monitors (Fig. 2a,b).Fig. 2: scRNA-seq characterization of candidate genes.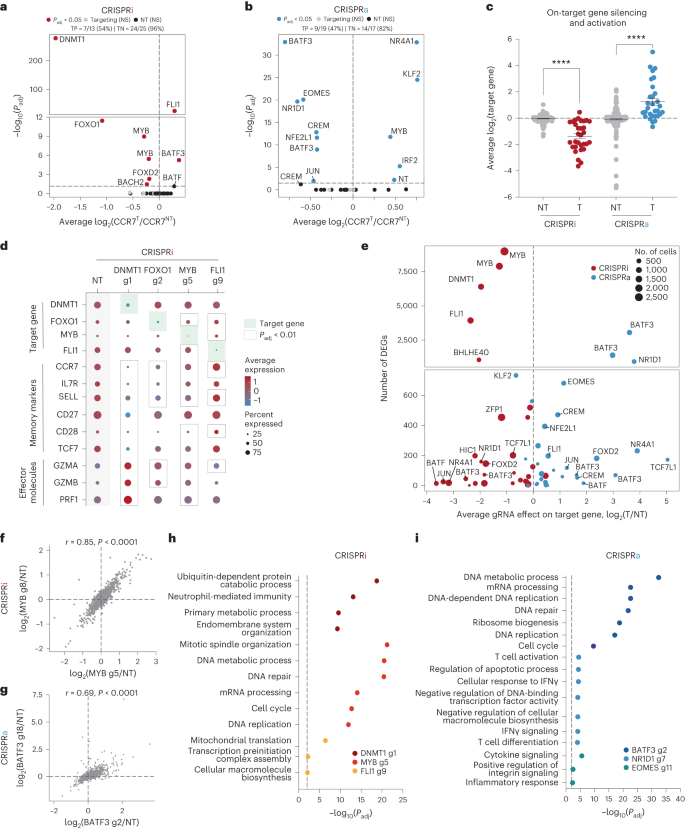 a,b, Importance (Padj) as opposed to reasonable fold exchange of CCR7 expression for each and every gRNA in comparison to nonperturbed cells for CRISPRi (a) and CRISPRa (b) perturbations. Vital gRNA results on CCR7 expression have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. True certain (TP) and adverse charges (TN) are displayed above each and every volcano plot. c, Fold exchange in goal gene expression for NT gRNAs and concentrating on gRNAs throughout CRISPRi (n = 31 gRNAs) and CRISPRa (n = 30 gRNAs) perturbations (imply values ± s.e.m.). A two-way ANOVA with Tukey’s put up hoc examine used to be used to match teams. d, Dot plot with reasonable expression and share of cells expressing goal genes, reminiscence markers and effector molecules for the indicated CRISPRi perturbations. Vital gRNA–gene hyperlinks have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. e, Collection of DEGs (Padj < 0.01) related to each and every gRNA as opposed to the gRNA impact at the goal gene for each CRISPRi and CRISPRa perturbations. f,g, Vital gRNA–gene hyperlinks have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. Correlation of the union set of DEGs between the highest two CRISPRi MYB gRNAs (f) and CRISPRa BATF3 gRNAs (g). Pearson’s correlation coefficient used to be calculated after which a two-tailed t-test used to be carried out to resolve whether or not the connection used to be vital. h,i, Consultant enriched pathways for the highest 3 CRISPRi (h) and CRISPRa gRNAs (i). Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction.We subsequent measured on-target gene silencing or activation. Of gRNAs assigned to no less than 5 cells in each and every of the CRISPRa and CRISPRi monitors, 56/61 gRNAs (92%) silenced or activated their gene goal (Fig. 2c). For the reason that CCR7 used to be decided on as a surrogate marker for a reminiscence T mobile phenotype, we anticipated some perturbations to control subset-defining gene expression techniques. Certainly, scRNA-seq printed that silencing the highest predicted certain regulators of reminiscence (DNMT1, FOXO1 and MYB) resulted in diminished expression of CCR7 and different memory-associated genes (similar to IL7R, SELL, CD27, CD28 and TCF7) and larger expression of effector-associated genes (GZMA, GZMB and PRF1) (Fig. 2nd).In spite of everything, we tested all differentially expressed genes (DEGs) related to each and every perturbation. Endogenous law of a number of TFs and epigenetic-modifying proteins had popular transcriptional results with six gene perturbations (4 CRISPRi gene perturbations and two CRISPRa gene perturbations) changing expression of >1,000 genes (Fig. 2e). Apparently, MYB repression with two distinctive gRNAs resulted in popular and concordant gene expression adjustments with 8,976 and seven,899 DEGs (Fig. 2e,f). MYB silencing drove a transcriptional program with hallmark options of effector T cells, suggesting that MYB performs a key function in regulating T mobile stemness in human CD8+ T cells as prior to now reported in mouse CD8+ T cells33 (Prolonged Information Fig. 4a,b and Supplementary Word 5).Endogenous activation of a number of TFs together with NR1D1, EOMES and BATF3 had pronounced results on T mobile state (Fig. 2e,i). Perturbation-driven single-cell clustering printed a definite mobile cluster with NR1D1 activation that used to be markedly enriched for exhaustion-associated genes in comparison to nonperturbed cells (Prolonged Information Fig. 4c–e and Supplementary Word 6). Significantly, a couple of extremely concordant BATF3 gRNAs had the most powerful results amongst CRISPRa perturbations with 3,056 and 1,402 DEGs (Fig. 2e,g). Gene Ontology analyses printed that BATF3-induced genes have been enriched for DNA and messenger RNA metabolic processing, ribosomal biogenesis and cell-cycle pathways, suggesting an growth in T mobile health (Fig. 2i).BATF3 OE techniques options of reminiscence T cellsBATF3 promotes survival and reminiscence formation in mouse CD8+ T cells. Then again, molecular and phenotypic results of BATF3 in human CD8+ T cells have now not been properly defined37. For the reason that BATF3 ORF supply led to better expression of BATF3 than endogenous BATF3 activation (Prolonged Information Fig. 5a and Supplementary Word 7) and the compact measurement of the BATF3 ORF (most effective 381 bp), we used ectopic BATF3 expression for all next assays and GFP OE as a adverse keep watch over.BATF3 OE markedly larger expression of IL7R, a floor marker related to T mobile survival, long-term endurance and certain medical reaction to ACT38 (Fig. 3a,b and Prolonged Information Fig. 5b). We carried out RNA-seq throughout CD8+ T cells from 5 donors to realize an impartial view of the transcriptomic adjustments brought on by means of BATF3 OE. In comparison to keep watch over cells, there have been over 1,100 DEGs dispensed nearly similarly between upregulated and downregulated genes (Fig. 3c). Gene Ontology analyses printed that BATF3 OE larger expression of genes excited by metabolic pathways similar to glycolysis and gluconeogenesis, DNA replication and translation (Fig. 3d and Supplementary Desk 4).Fig. 3: BATF3 OE promotes particular options of reminiscence T cells and counters exhaustion and cytotoxic gene signatures.
a,b, Importance (Padj) as opposed to reasonable fold exchange of CCR7 expression for each and every gRNA in comparison to nonperturbed cells for CRISPRi (a) and CRISPRa (b) perturbations. Vital gRNA results on CCR7 expression have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. True certain (TP) and adverse charges (TN) are displayed above each and every volcano plot. c, Fold exchange in goal gene expression for NT gRNAs and concentrating on gRNAs throughout CRISPRi (n = 31 gRNAs) and CRISPRa (n = 30 gRNAs) perturbations (imply values ± s.e.m.). A two-way ANOVA with Tukey’s put up hoc examine used to be used to match teams. d, Dot plot with reasonable expression and share of cells expressing goal genes, reminiscence markers and effector molecules for the indicated CRISPRi perturbations. Vital gRNA–gene hyperlinks have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. e, Collection of DEGs (Padj < 0.01) related to each and every gRNA as opposed to the gRNA impact at the goal gene for each CRISPRi and CRISPRa perturbations. f,g, Vital gRNA–gene hyperlinks have been outlined the usage of a two-tailed MAST examine with Bonferroni correction. Correlation of the union set of DEGs between the highest two CRISPRi MYB gRNAs (f) and CRISPRa BATF3 gRNAs (g). Pearson’s correlation coefficient used to be calculated after which a two-tailed t-test used to be carried out to resolve whether or not the connection used to be vital. h,i, Consultant enriched pathways for the highest 3 CRISPRi (h) and CRISPRa gRNAs (i). Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction.We subsequent measured on-target gene silencing or activation. Of gRNAs assigned to no less than 5 cells in each and every of the CRISPRa and CRISPRi monitors, 56/61 gRNAs (92%) silenced or activated their gene goal (Fig. 2c). For the reason that CCR7 used to be decided on as a surrogate marker for a reminiscence T mobile phenotype, we anticipated some perturbations to control subset-defining gene expression techniques. Certainly, scRNA-seq printed that silencing the highest predicted certain regulators of reminiscence (DNMT1, FOXO1 and MYB) resulted in diminished expression of CCR7 and different memory-associated genes (similar to IL7R, SELL, CD27, CD28 and TCF7) and larger expression of effector-associated genes (GZMA, GZMB and PRF1) (Fig. 2nd).In spite of everything, we tested all differentially expressed genes (DEGs) related to each and every perturbation. Endogenous law of a number of TFs and epigenetic-modifying proteins had popular transcriptional results with six gene perturbations (4 CRISPRi gene perturbations and two CRISPRa gene perturbations) changing expression of >1,000 genes (Fig. 2e). Apparently, MYB repression with two distinctive gRNAs resulted in popular and concordant gene expression adjustments with 8,976 and seven,899 DEGs (Fig. 2e,f). MYB silencing drove a transcriptional program with hallmark options of effector T cells, suggesting that MYB performs a key function in regulating T mobile stemness in human CD8+ T cells as prior to now reported in mouse CD8+ T cells33 (Prolonged Information Fig. 4a,b and Supplementary Word 5).Endogenous activation of a number of TFs together with NR1D1, EOMES and BATF3 had pronounced results on T mobile state (Fig. 2e,i). Perturbation-driven single-cell clustering printed a definite mobile cluster with NR1D1 activation that used to be markedly enriched for exhaustion-associated genes in comparison to nonperturbed cells (Prolonged Information Fig. 4c–e and Supplementary Word 6). Significantly, a couple of extremely concordant BATF3 gRNAs had the most powerful results amongst CRISPRa perturbations with 3,056 and 1,402 DEGs (Fig. 2e,g). Gene Ontology analyses printed that BATF3-induced genes have been enriched for DNA and messenger RNA metabolic processing, ribosomal biogenesis and cell-cycle pathways, suggesting an growth in T mobile health (Fig. 2i).BATF3 OE techniques options of reminiscence T cellsBATF3 promotes survival and reminiscence formation in mouse CD8+ T cells. Then again, molecular and phenotypic results of BATF3 in human CD8+ T cells have now not been properly defined37. For the reason that BATF3 ORF supply led to better expression of BATF3 than endogenous BATF3 activation (Prolonged Information Fig. 5a and Supplementary Word 7) and the compact measurement of the BATF3 ORF (most effective 381 bp), we used ectopic BATF3 expression for all next assays and GFP OE as a adverse keep watch over.BATF3 OE markedly larger expression of IL7R, a floor marker related to T mobile survival, long-term endurance and certain medical reaction to ACT38 (Fig. 3a,b and Prolonged Information Fig. 5b). We carried out RNA-seq throughout CD8+ T cells from 5 donors to realize an impartial view of the transcriptomic adjustments brought on by means of BATF3 OE. In comparison to keep watch over cells, there have been over 1,100 DEGs dispensed nearly similarly between upregulated and downregulated genes (Fig. 3c). Gene Ontology analyses printed that BATF3 OE larger expression of genes excited by metabolic pathways similar to glycolysis and gluconeogenesis, DNA replication and translation (Fig. 3d and Supplementary Desk 4).Fig. 3: BATF3 OE promotes particular options of reminiscence T cells and counters exhaustion and cytotoxic gene signatures.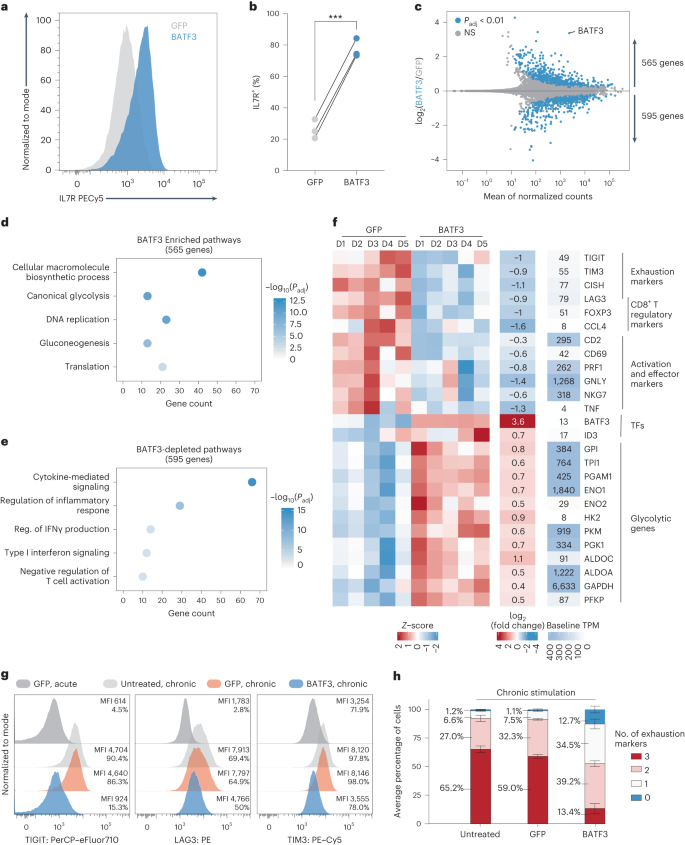 a, Consultant histogram of IL7R expression in CD8+ T cells with BATF3 OE or keep watch over GFP OE on day 8 post-transduction. b, Abstract statistics of IL7R expression without or with BATF3 OE (n = 3 donors with traces connecting the similar donor, a two-tailed paired t-test (P = 0.0004) used to be used to match IL7R expression between teams). c, Differential gene expression research between CD8+ T cells without or with BATF3 OE on day 10 put up transduction (n = 5 donors). DEGs have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. d,e, Decided on enriched (d) and depleted (e) organic processes from BATF3 OE. Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction. f, Heatmap of DEGs (Padj < 0.01, n = 5 donors) associated with T mobile exhaustion, regulatory operate, cytotoxicity, transcriptional task and glycolysis. g, Consultant histograms of exhaustion markers (TIGIT, LAG3 and TIM3) on day 12 after acute or power stimulation throughout teams. h, Stacked bar chart with reasonable share of CD8+ T cells certain for 0, one, two or 3 exhaustion markers (TIGIT, LAG3 and TIM3) on day 12 after power stimulation throughout teams (n = 3 donors, imply values ± s.e.m.).Against this, BATF3 OE dampened T mobile effector techniques and downregulated activation markers, inflammatory cytokines and cytotoxic molecules (Fig. 3e,f). Moreover, BATF3 OE lowered expression of a number of markers related to FOXP3+ regulatory T cells (Tregs), which might be related to deficient reaction to ACT38. A subset of CD8+FOXP3+LAG3+ Tregs suppress T mobile task by means of secreting CC chemokine ligand 4 (CCL4)39. Apparently, BATF3 OE lowered expression of FOXP3, LAG3 and CCL4 in CD8+ T cells (Fig. 3f and Prolonged Information Fig. 5c).Along with LAG3, BATF3 silenced different canonical markers of T mobile exhaustion together with TIGIT, TIM3 and CISH (Fig. 3f and Prolonged Information Fig. 5c). We speculated those results could be amplified within the context of power antigen stimulation (Prolonged Information Fig. 6). As prior to now observed40, PD1 expression peaked after the preliminary stimulation after which tapered off over the years, while TIGIT, LAG3 and TIM3 expression used to be maintained or larger after each and every next spherical of stimulation. Significantly, BATF3 OE attenuated PD1 induction and limited TIGIT, LAG3 and TIM3 expression to intently resemble that of acutely stimulated cells in spite of 3 further rounds of TCR stimulation (Fig. 3g and Prolonged Information Fig. 6b,c). As terminally exhausted T cells frequently co-express more than one exhaustion-associated markers, we quantified the share of cells expressing each and every mixture of TIGIT, LAG3 and TIM3. Simplest 13% of BATF3 OE T cells co-expressed all 3 markers in comparison to 65% and 59% of untreated and GFP-treated T cells (Fig. 3h).BATF3 OE remodels the epigenetic landscapeAs an orthogonal manner of inducing T mobile exhaustion, we acutely or chronically stimulated HER2-targeted CAR T cells without or with BATF3 OE with HER2+ most cancers cells (Fig. 4, Supplementary Fig. 8 and Supplementary Word 8). We assessed chromatin reworking by means of assay for transposase-accessible chromatin with sequencing (ATAC-seq) in line with BATF3 OE below acute or power stimulation. In each fashions, BATF3 OE widely made over the chromatin with 5,104 and 22,201 differentially out there areas in comparison to keep watch over T cells with 60% and 54% of those areas, respectively, being extra out there with BATF3 OE (Fig. 4a–c). A lot of these adjustments have been in intronic or intergenic areas in keeping with cis-regulatory or enhancer parts (Prolonged Information Fig. 7a,b).Fig. 4: BATF3 OE remodels the chromatin panorama within the context of acute or power T mobile stimulation.
a, Consultant histogram of IL7R expression in CD8+ T cells with BATF3 OE or keep watch over GFP OE on day 8 post-transduction. b, Abstract statistics of IL7R expression without or with BATF3 OE (n = 3 donors with traces connecting the similar donor, a two-tailed paired t-test (P = 0.0004) used to be used to match IL7R expression between teams). c, Differential gene expression research between CD8+ T cells without or with BATF3 OE on day 10 put up transduction (n = 5 donors). DEGs have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. d,e, Decided on enriched (d) and depleted (e) organic processes from BATF3 OE. Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction. f, Heatmap of DEGs (Padj < 0.01, n = 5 donors) associated with T mobile exhaustion, regulatory operate, cytotoxicity, transcriptional task and glycolysis. g, Consultant histograms of exhaustion markers (TIGIT, LAG3 and TIM3) on day 12 after acute or power stimulation throughout teams. h, Stacked bar chart with reasonable share of CD8+ T cells certain for 0, one, two or 3 exhaustion markers (TIGIT, LAG3 and TIM3) on day 12 after power stimulation throughout teams (n = 3 donors, imply values ± s.e.m.).Against this, BATF3 OE dampened T mobile effector techniques and downregulated activation markers, inflammatory cytokines and cytotoxic molecules (Fig. 3e,f). Moreover, BATF3 OE lowered expression of a number of markers related to FOXP3+ regulatory T cells (Tregs), which might be related to deficient reaction to ACT38. A subset of CD8+FOXP3+LAG3+ Tregs suppress T mobile task by means of secreting CC chemokine ligand 4 (CCL4)39. Apparently, BATF3 OE lowered expression of FOXP3, LAG3 and CCL4 in CD8+ T cells (Fig. 3f and Prolonged Information Fig. 5c).Along with LAG3, BATF3 silenced different canonical markers of T mobile exhaustion together with TIGIT, TIM3 and CISH (Fig. 3f and Prolonged Information Fig. 5c). We speculated those results could be amplified within the context of power antigen stimulation (Prolonged Information Fig. 6). As prior to now observed40, PD1 expression peaked after the preliminary stimulation after which tapered off over the years, while TIGIT, LAG3 and TIM3 expression used to be maintained or larger after each and every next spherical of stimulation. Significantly, BATF3 OE attenuated PD1 induction and limited TIGIT, LAG3 and TIM3 expression to intently resemble that of acutely stimulated cells in spite of 3 further rounds of TCR stimulation (Fig. 3g and Prolonged Information Fig. 6b,c). As terminally exhausted T cells frequently co-express more than one exhaustion-associated markers, we quantified the share of cells expressing each and every mixture of TIGIT, LAG3 and TIM3. Simplest 13% of BATF3 OE T cells co-expressed all 3 markers in comparison to 65% and 59% of untreated and GFP-treated T cells (Fig. 3h).BATF3 OE remodels the epigenetic landscapeAs an orthogonal manner of inducing T mobile exhaustion, we acutely or chronically stimulated HER2-targeted CAR T cells without or with BATF3 OE with HER2+ most cancers cells (Fig. 4, Supplementary Fig. 8 and Supplementary Word 8). We assessed chromatin reworking by means of assay for transposase-accessible chromatin with sequencing (ATAC-seq) in line with BATF3 OE below acute or power stimulation. In each fashions, BATF3 OE widely made over the chromatin with 5,104 and 22,201 differentially out there areas in comparison to keep watch over T cells with 60% and 54% of those areas, respectively, being extra out there with BATF3 OE (Fig. 4a–c). A lot of these adjustments have been in intronic or intergenic areas in keeping with cis-regulatory or enhancer parts (Prolonged Information Fig. 7a,b).Fig. 4: BATF3 OE remodels the chromatin panorama within the context of acute or power T mobile stimulation.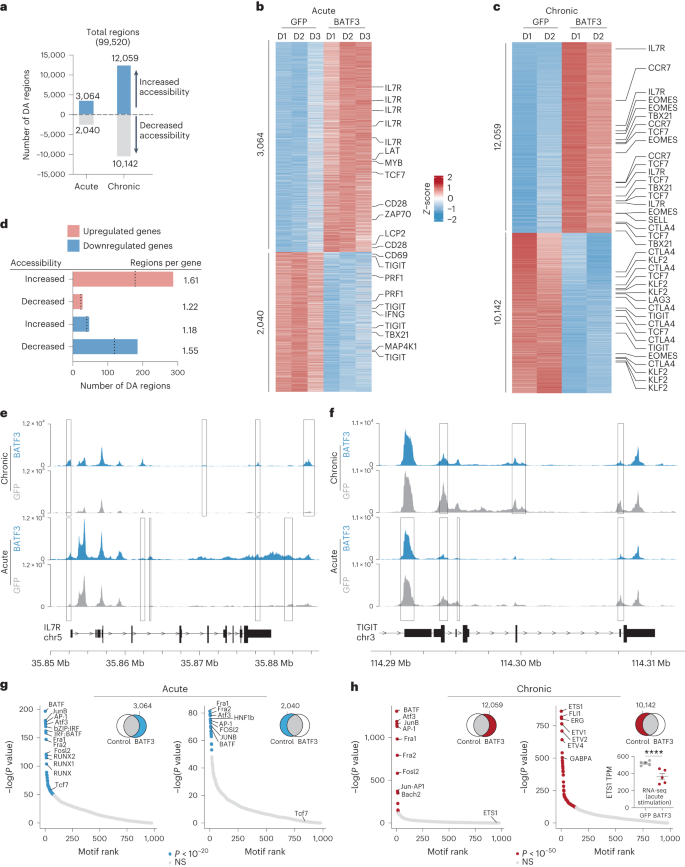 a, Collection of ATAC-seq areas with larger or diminished accessibility in acutely (n = 3 donors) or chronically stimulated CD8+ T cells (n = 2 donors) with BATF3 OE on day 14 post-transduction. Differentially out there (DA) areas have been outlined as Padj < 0.05 the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. b,c, Heatmap of DA areas between keep watch over and BATF3 OE T cells below acute (b) or power (c) stimulation with decided on areas annotated with their nearest gene. d, Joint research of RNA-seq and ATAC-seq datasets within the context of acute stimulation. Collection of DA areas close to upregulated and downregulated genes. Dashed traces constitute the choice of distinctive DEGs related to DA areas. e,f, Consultant ATAC-seq tracks of IL7R (e) and TIGIT (f) loci after acute or power stimulation with overlaid rectangles indicating DA areas between keep watch over and BATF3 OE T cells in each and every context. g,h, TF DNA-binding motifs enriched in open (left) and closed (proper) areas of chromatin in BATF3 OE T cells in comparison to keep watch over T cells after acute (g) and persistent (h) stimulation. HOMER computes P values from the cumulative hypergeometric distribution and does now not modify for more than one hypotheses. Bar plot in decrease proper nook illustrates BATF3’s impact on ETS1 expression in keeping with RNA-seq (n = 5 donors, imply values ± s.e.m.; statistical importance used to be decided the usage of a paired two-tailed DESeq2 examine between medication teams).To know whether or not adjustments in chromatin accessibility corresponded to adjustments in gene expression, we collectively analyzed our ATAC-seq and RNA-seq knowledge within the context of acute stimulation. We assigned each and every differentially out there area to its closest gene to estimate genes which may be regulated in cis by means of those parts. There used to be an enrichment of areas with larger or diminished accessibility proximal to upregulated and downregulated genes, respectively, indicating that BATF3-driven epigenetic adjustments affected close by gene transcription (Fig. 4d). Roughly 25% of the genes that modified expression have been related to a corresponding differentially out there area (297 out of one,160 genes). As an example, BATF3 OE larger accessibility on the IL7R promoter, intronic, 3′-untranslated area, and intergenic areas and diminished accessibility on the 5′-untranslated area, intronic and exonic areas of TIGIT (Fig. 4d,e). Moreover, BATF3 OE in part counteracted the impact of power antigen stimulation at each and every of those loci (Fig. 4d,e). Apparently, BATF3 OE larger accessibility at areas close to each reminiscence (TCF7, MYB, IL7R, CCR7 and SELL) and effector-associated genes (EOMES and TBX21) (Fig. 4c). This will likely constitute a hybrid T mobile phenotype or the presence of heterogenous subpopulations of reminiscence and effector T cells. In line with RNA-seq and go with the flow knowledge, there used to be lowered accessibility at exhaustion-associated loci similar to TIGIT, CTLA4 and LAG3 with BATF3 OE.Subsequent, we carried out motif enrichment analyses to realize additional perception into the transcriptional networks regulating keep watch over and BATF3 OE T cells below acute and persistent stimulation (Fig. 4g,h). In comparison to keep watch over T cells, AP-1 transcription circle of relatives motifs have been strongly enriched in each differentially open and closed areas with BATF3 OE below acute stimulation. In reality, 45% and 42% of differentially open and closed areas websites, respectively, harbored a BATF3 motif, suggesting direct BATF3 task at those areas. That is in keeping with the twin attainable of BATF3 to silence or turn on gene expression relying on its binding partners41. Apparently, a TCF7 binding motif used to be uniquely enriched in differentially open areas with BATF3 OE. Then again, below power stimulation, AP-1 TF motifs have been enriched with BATF3 OE most effective in differentially open areas. ETS circle of relatives member motifs have been enriched in closed areas, suggesting that BATF3 OE dampens the task of those elements. A number of ETS members of the family (for instance ETV1, ETV2 and ETV4) don’t seem to be expressed at baseline in T cells, making it not going those genes give a contribution to the popular epigenetic adjustments brought on by means of power antigen stimulation. ETS1, then again, would possibly constitute a very powerful node of the transcriptional community as it’s extremely expressed at baseline (>500 transcripts according to million, TPM) and considerably repressed by means of BATF3 OE below acute stimulation (Fig. 4h).BATF3 OE complements efficiency of CAR T cellsGiven the profound transcriptional and epigenetic adjustments, we hypothesized that BATF3 OE may make stronger CD8+ T mobile operate. First, we noticed that BATF3 OE larger killing of cultured human HER2+ most cancers cells by means of HER2-targeted CAR T cells throughout donors and effector:goal (E:T) ratios (Fig. 5a and Prolonged Information Fig. 8a,b). Subsequent, we evaluated whether or not BATF3 OE may just make stronger in vivo keep watch over of cast tumors, given the problem of T mobile exhaustion within the cast tumor setting42,43. To simplify supply of the CAR and BATF3 transgenes, we built all-in-one lentiviral vectors encoding a HER2 CAR coupled to both GFP or BATF3 expression. Strikingly, CAR T cells co-expressing BATF3 markedly enhanced tumor keep watch over at two subcurative doses (2.5 × 105 and 5 × 105 CAR+ cells) in comparison to keep watch over CAR T cells in an orthotopic human HER2+ breast most cancers fashion (Fig. 5b,c and Prolonged Information Fig. 8c–f).Fig. 5: BATF3 OE complements CAR T mobile efficiency.
a, Collection of ATAC-seq areas with larger or diminished accessibility in acutely (n = 3 donors) or chronically stimulated CD8+ T cells (n = 2 donors) with BATF3 OE on day 14 post-transduction. Differentially out there (DA) areas have been outlined as Padj < 0.05 the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. b,c, Heatmap of DA areas between keep watch over and BATF3 OE T cells below acute (b) or power (c) stimulation with decided on areas annotated with their nearest gene. d, Joint research of RNA-seq and ATAC-seq datasets within the context of acute stimulation. Collection of DA areas close to upregulated and downregulated genes. Dashed traces constitute the choice of distinctive DEGs related to DA areas. e,f, Consultant ATAC-seq tracks of IL7R (e) and TIGIT (f) loci after acute or power stimulation with overlaid rectangles indicating DA areas between keep watch over and BATF3 OE T cells in each and every context. g,h, TF DNA-binding motifs enriched in open (left) and closed (proper) areas of chromatin in BATF3 OE T cells in comparison to keep watch over T cells after acute (g) and persistent (h) stimulation. HOMER computes P values from the cumulative hypergeometric distribution and does now not modify for more than one hypotheses. Bar plot in decrease proper nook illustrates BATF3’s impact on ETS1 expression in keeping with RNA-seq (n = 5 donors, imply values ± s.e.m.; statistical importance used to be decided the usage of a paired two-tailed DESeq2 examine between medication teams).To know whether or not adjustments in chromatin accessibility corresponded to adjustments in gene expression, we collectively analyzed our ATAC-seq and RNA-seq knowledge within the context of acute stimulation. We assigned each and every differentially out there area to its closest gene to estimate genes which may be regulated in cis by means of those parts. There used to be an enrichment of areas with larger or diminished accessibility proximal to upregulated and downregulated genes, respectively, indicating that BATF3-driven epigenetic adjustments affected close by gene transcription (Fig. 4d). Roughly 25% of the genes that modified expression have been related to a corresponding differentially out there area (297 out of one,160 genes). As an example, BATF3 OE larger accessibility on the IL7R promoter, intronic, 3′-untranslated area, and intergenic areas and diminished accessibility on the 5′-untranslated area, intronic and exonic areas of TIGIT (Fig. 4d,e). Moreover, BATF3 OE in part counteracted the impact of power antigen stimulation at each and every of those loci (Fig. 4d,e). Apparently, BATF3 OE larger accessibility at areas close to each reminiscence (TCF7, MYB, IL7R, CCR7 and SELL) and effector-associated genes (EOMES and TBX21) (Fig. 4c). This will likely constitute a hybrid T mobile phenotype or the presence of heterogenous subpopulations of reminiscence and effector T cells. In line with RNA-seq and go with the flow knowledge, there used to be lowered accessibility at exhaustion-associated loci similar to TIGIT, CTLA4 and LAG3 with BATF3 OE.Subsequent, we carried out motif enrichment analyses to realize additional perception into the transcriptional networks regulating keep watch over and BATF3 OE T cells below acute and persistent stimulation (Fig. 4g,h). In comparison to keep watch over T cells, AP-1 transcription circle of relatives motifs have been strongly enriched in each differentially open and closed areas with BATF3 OE below acute stimulation. In reality, 45% and 42% of differentially open and closed areas websites, respectively, harbored a BATF3 motif, suggesting direct BATF3 task at those areas. That is in keeping with the twin attainable of BATF3 to silence or turn on gene expression relying on its binding partners41. Apparently, a TCF7 binding motif used to be uniquely enriched in differentially open areas with BATF3 OE. Then again, below power stimulation, AP-1 TF motifs have been enriched with BATF3 OE most effective in differentially open areas. ETS circle of relatives member motifs have been enriched in closed areas, suggesting that BATF3 OE dampens the task of those elements. A number of ETS members of the family (for instance ETV1, ETV2 and ETV4) don’t seem to be expressed at baseline in T cells, making it not going those genes give a contribution to the popular epigenetic adjustments brought on by means of power antigen stimulation. ETS1, then again, would possibly constitute a very powerful node of the transcriptional community as it’s extremely expressed at baseline (>500 transcripts according to million, TPM) and considerably repressed by means of BATF3 OE below acute stimulation (Fig. 4h).BATF3 OE complements efficiency of CAR T cellsGiven the profound transcriptional and epigenetic adjustments, we hypothesized that BATF3 OE may make stronger CD8+ T mobile operate. First, we noticed that BATF3 OE larger killing of cultured human HER2+ most cancers cells by means of HER2-targeted CAR T cells throughout donors and effector:goal (E:T) ratios (Fig. 5a and Prolonged Information Fig. 8a,b). Subsequent, we evaluated whether or not BATF3 OE may just make stronger in vivo keep watch over of cast tumors, given the problem of T mobile exhaustion within the cast tumor setting42,43. To simplify supply of the CAR and BATF3 transgenes, we built all-in-one lentiviral vectors encoding a HER2 CAR coupled to both GFP or BATF3 expression. Strikingly, CAR T cells co-expressing BATF3 markedly enhanced tumor keep watch over at two subcurative doses (2.5 × 105 and 5 × 105 CAR+ cells) in comparison to keep watch over CAR T cells in an orthotopic human HER2+ breast most cancers fashion (Fig. 5b,c and Prolonged Information Fig. 8c–f).Fig. 5: BATF3 OE complements CAR T mobile efficiency.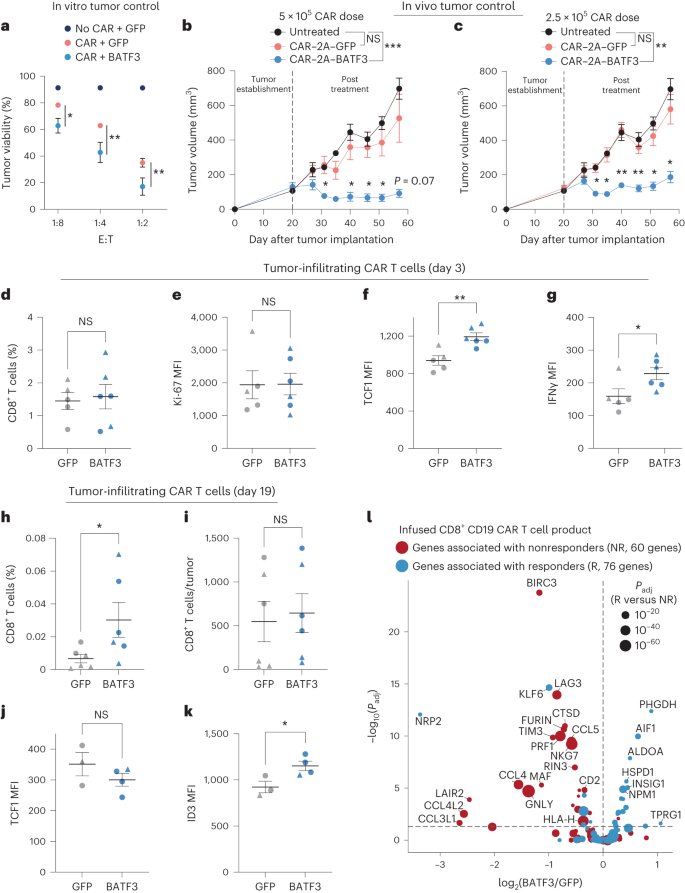 a, Tumor viability after co-culture at specified E:T ratios (n = 3 donors). A two-way ANOVA with Dunnett’s put up hoc examine when put next tumor viability at each and every E:T ratio: 1:8 (Padj = 0.0243), 1:4 (Padj = 0.0042) and 1:2 (Padj = 0.0099). b,c, Tumor volumes of untreated (n = 5) and handled mice with 5 × 105 (n = 1 donor, 5 mice according to medication) (b) or 2.5 × 105 CAR T cells (n = 1 donor, 4 mice according to medication) (c) without or with BATF3 OE. Two-way ANOVA with Tukey’s put up hoc checks when put next tumor volumes at each and every time level throughout remedies. Tumor volumes have been by no means other between untreated and keep watch over CAR teams. Asterisks point out vital variations between keep watch over and BATF3 OE CAR T cells. d–g, Proportion of CD8+ T cells (d) inside each and every resected tumor on day 3 post-treatment and (Ki-67 (e), TCF1 (f) and IFNγ (g) MFI of T cells (n = 2 donors, 2 GFP and three BATF3 mice for donor 1, 3 mice according to medication for donor 2). Two-tailed Mann–Whitney checks when put next share of CD8+ cells and marker MFI between teams (P = 0.0065 for TCF1 and P = 0.0303 for IFNγ). h,i, Proportion (h) and overall quantity (i) of CD8+ T cells inside each and every resected tumor on day 19 post-treatment (n = 2 donors, 4 mice according to medication for donor 1, 2 GFP and three BATF3 mice for donor 2). Two-tailed Mann–Whitney checks when put next share (P = 0.026) and overall choice of CD8+ cells between teams. j,ok, TCF1 and ID3 MFI of T cells on day 19 (n = 2 donors, 1 mouse according to medication for donor 1, 2 GFP and three BATF3 mice for donor 2). Two-tailed t-tests when put next MFI between teams (P = 0.037 for ID3). l, Importance (Padj) as opposed to fold exchange between BATF3 OE and keep watch over CD8+ T cells for 144 genes related to medical result to CD19 CAR T mobile therapy38. Imply values ± s.e.m. are plotted for a–ok.To discover the mechanism riding awesome tumor keep watch over with BATF3 OE, we repeated the in vivo experiment with T cells from two other donors and phenotypically characterised the CAR T cells ahead of medication and after gathering tumor-infiltrating CAR T cells on day 3 and day 19 post-treatment (Fig. 5d–ok, Prolonged Information Fig. 9 and Supplementary Fig. 9). Throughout each units of experiments, there have been no variations in CAR transduction charges (>70% for all teams) or the overall choice of CAR+ T cells ahead of intravenous injections between CAR constructs (Prolonged Information Fig. 8d,e). Once more, we noticed awesome tumor keep watch over with BATF3 OE CAR T cells throughout each donors (Prolonged Information Fig. 9a,b). In line with the former characterization (Fig. 3f–h), enter BATF3 OE cells tended to specific decrease ranges of exhaustion markers together with LAG3, TIGIT and TIM3 (Prolonged Information Fig. 9c).Extra hanging variations between the 2 teams emerged on the day 3 post-treatment time level. In keep watch over and BATF3 OE cells, we detected identical proportions of CD8+ T cells inside the tumor and circulating in peripheral blood, indicating that BATF3 OE used to be now not making improvements to tumor keep watch over by means of simply expanding T mobile proliferation or tumor trafficking (Fig. 5d and Prolonged Information Fig. 9f). In a similar fashion, expression of the proliferative marker Ki-67 used to be identical between the teams (Fig. 5e). Reasonably, we spotted that tumor-infiltrating CAR T cells with BATF3 OE expressed upper ranges of each TCF1 and IFNγ (Fig. 5f,g). This caused us to revisit our gene expression and chromatin accessibility knowledge. BATF3 OE didn’t build up expression of TCF7 (which encodes for TCF1) below acute stimulation (Prolonged Information Fig. 5c). Then again, there have been seven differentially out there websites close to the TCF7 locus between keep watch over and BATF3 OE CAR T cells below power stimulation (Fig. 4c and Prolonged Information Fig. 7c). Significantly, 5/7 websites have been extra out there in BATF3 OE cells together with all 3 intragenic areas. Those knowledge recommend that BATF3 OE can in part counter heterochromatinization of the TCF7 locus throughout power antigen stimulation and retain upper ranges of TCF1 expression.As mirrored within the tumor enlargement curves, we detected a better share of tumor-infiltrating CAR T cells within the BATF3 OE staff on the ultimate day 19 time level, most likely because of smaller tumor sizes, as absolutely the choice of T cells have been identical between the 2 teams (Fig. 5h,i). We didn’t stumble on any CAR T cells in peripheral blood for both staff. We stained the tumor-infiltrating CAR T cells for TCF1, TBET, EOMES, GATA3, ID2, ID3 and IRF4. Apparently, TCF1 used to be now not differentially expressed, however ID3 (a downstream TF of TCF1 (ref. 44)) used to be upregulated within the BATF3 OE staff (Fig. 5j,ok). Subsequently, BATF3 OE T cells could have steadily transitioned from transcriptional techniques pushed by means of TCF1 to ID3.Given the improved tumor keep watch over conferred by means of BATF3 OE in CD8+ T cells, we investigated whether or not BATF3 OE programmed a transcriptional signature related to medical reaction to ACT. In reality, nonresponders to CD19-targeted CAR T mobile treatment had a considerably upper share of CD8+ T cells in a cytotoxic or exhausted phenotype in comparison to responders in a up to date medical trial38. The usage of those datasets, we recognized 147 DEGs between the infused CD8+ CAR T mobile fabricated from responders and nonresponders (Supplementary Fig. 10). Of those 147 DEGs, 144 genes have been detected in our RNA-seq knowledge. Strikingly, BATF3 OE silenced 35% (23/65) of genes related to nonresponse and activated 20% (16/79) of genes related to reaction (Fig. 5l). Seven of the 10 genes maximum strongly related to medical result have been regulated in a positive route. Conversely, most effective 4.9% (7/144) of genes have been regulated in a route opposing certain medical reaction, offering additional proof that BATF3 OE drives a transcriptional program related to certain medical results.CRISPRko monitors expose cofactors of BATF3BATF3 is a compact AP-1 TF with just a fundamental DNA binding area and a leucine zipper motif. For the reason that BATF3 lacks further protein domain names similar to transactivation domain names for gene activation, we speculated that BATF3 interacts with different TFs to have an effect on gene expression and chromatin accessibility (Supplementary Word 9)41. Moreover, we reasoned that different TFs may compete with or inhibit BATF3 and that disposing of those elements would additional enlarge the results of BATF3 OE. To spot those elements, we carried out parallel CRISPRko monitors without or with BATF3 OE the usage of a gRNA library concentrating on all 1,612 human TF genes45 (TFome) (Fig. 6a). We decided on IL7R expression because the readout for those monitors as a result of BATF3 OE profoundly will increase IL7R expression (Fig. 3a,b), thus offering a proxy for BATF3 task. IL7R could also be expressed in 20–50% of CD8+ T cells at baseline, making it possible to recuperate gene hits in each instructions, in contrast to ubiquitously silenced and extremely expressed genes.Fig. 6: CRISPRko monitors expose cofactors of BATF3 and different objectives for most cancers immunotherapy.
a, Tumor viability after co-culture at specified E:T ratios (n = 3 donors). A two-way ANOVA with Dunnett’s put up hoc examine when put next tumor viability at each and every E:T ratio: 1:8 (Padj = 0.0243), 1:4 (Padj = 0.0042) and 1:2 (Padj = 0.0099). b,c, Tumor volumes of untreated (n = 5) and handled mice with 5 × 105 (n = 1 donor, 5 mice according to medication) (b) or 2.5 × 105 CAR T cells (n = 1 donor, 4 mice according to medication) (c) without or with BATF3 OE. Two-way ANOVA with Tukey’s put up hoc checks when put next tumor volumes at each and every time level throughout remedies. Tumor volumes have been by no means other between untreated and keep watch over CAR teams. Asterisks point out vital variations between keep watch over and BATF3 OE CAR T cells. d–g, Proportion of CD8+ T cells (d) inside each and every resected tumor on day 3 post-treatment and (Ki-67 (e), TCF1 (f) and IFNγ (g) MFI of T cells (n = 2 donors, 2 GFP and three BATF3 mice for donor 1, 3 mice according to medication for donor 2). Two-tailed Mann–Whitney checks when put next share of CD8+ cells and marker MFI between teams (P = 0.0065 for TCF1 and P = 0.0303 for IFNγ). h,i, Proportion (h) and overall quantity (i) of CD8+ T cells inside each and every resected tumor on day 19 post-treatment (n = 2 donors, 4 mice according to medication for donor 1, 2 GFP and three BATF3 mice for donor 2). Two-tailed Mann–Whitney checks when put next share (P = 0.026) and overall choice of CD8+ cells between teams. j,ok, TCF1 and ID3 MFI of T cells on day 19 (n = 2 donors, 1 mouse according to medication for donor 1, 2 GFP and three BATF3 mice for donor 2). Two-tailed t-tests when put next MFI between teams (P = 0.037 for ID3). l, Importance (Padj) as opposed to fold exchange between BATF3 OE and keep watch over CD8+ T cells for 144 genes related to medical result to CD19 CAR T mobile therapy38. Imply values ± s.e.m. are plotted for a–ok.To discover the mechanism riding awesome tumor keep watch over with BATF3 OE, we repeated the in vivo experiment with T cells from two other donors and phenotypically characterised the CAR T cells ahead of medication and after gathering tumor-infiltrating CAR T cells on day 3 and day 19 post-treatment (Fig. 5d–ok, Prolonged Information Fig. 9 and Supplementary Fig. 9). Throughout each units of experiments, there have been no variations in CAR transduction charges (>70% for all teams) or the overall choice of CAR+ T cells ahead of intravenous injections between CAR constructs (Prolonged Information Fig. 8d,e). Once more, we noticed awesome tumor keep watch over with BATF3 OE CAR T cells throughout each donors (Prolonged Information Fig. 9a,b). In line with the former characterization (Fig. 3f–h), enter BATF3 OE cells tended to specific decrease ranges of exhaustion markers together with LAG3, TIGIT and TIM3 (Prolonged Information Fig. 9c).Extra hanging variations between the 2 teams emerged on the day 3 post-treatment time level. In keep watch over and BATF3 OE cells, we detected identical proportions of CD8+ T cells inside the tumor and circulating in peripheral blood, indicating that BATF3 OE used to be now not making improvements to tumor keep watch over by means of simply expanding T mobile proliferation or tumor trafficking (Fig. 5d and Prolonged Information Fig. 9f). In a similar fashion, expression of the proliferative marker Ki-67 used to be identical between the teams (Fig. 5e). Reasonably, we spotted that tumor-infiltrating CAR T cells with BATF3 OE expressed upper ranges of each TCF1 and IFNγ (Fig. 5f,g). This caused us to revisit our gene expression and chromatin accessibility knowledge. BATF3 OE didn’t build up expression of TCF7 (which encodes for TCF1) below acute stimulation (Prolonged Information Fig. 5c). Then again, there have been seven differentially out there websites close to the TCF7 locus between keep watch over and BATF3 OE CAR T cells below power stimulation (Fig. 4c and Prolonged Information Fig. 7c). Significantly, 5/7 websites have been extra out there in BATF3 OE cells together with all 3 intragenic areas. Those knowledge recommend that BATF3 OE can in part counter heterochromatinization of the TCF7 locus throughout power antigen stimulation and retain upper ranges of TCF1 expression.As mirrored within the tumor enlargement curves, we detected a better share of tumor-infiltrating CAR T cells within the BATF3 OE staff on the ultimate day 19 time level, most likely because of smaller tumor sizes, as absolutely the choice of T cells have been identical between the 2 teams (Fig. 5h,i). We didn’t stumble on any CAR T cells in peripheral blood for both staff. We stained the tumor-infiltrating CAR T cells for TCF1, TBET, EOMES, GATA3, ID2, ID3 and IRF4. Apparently, TCF1 used to be now not differentially expressed, however ID3 (a downstream TF of TCF1 (ref. 44)) used to be upregulated within the BATF3 OE staff (Fig. 5j,ok). Subsequently, BATF3 OE T cells could have steadily transitioned from transcriptional techniques pushed by means of TCF1 to ID3.Given the improved tumor keep watch over conferred by means of BATF3 OE in CD8+ T cells, we investigated whether or not BATF3 OE programmed a transcriptional signature related to medical reaction to ACT. In reality, nonresponders to CD19-targeted CAR T mobile treatment had a considerably upper share of CD8+ T cells in a cytotoxic or exhausted phenotype in comparison to responders in a up to date medical trial38. The usage of those datasets, we recognized 147 DEGs between the infused CD8+ CAR T mobile fabricated from responders and nonresponders (Supplementary Fig. 10). Of those 147 DEGs, 144 genes have been detected in our RNA-seq knowledge. Strikingly, BATF3 OE silenced 35% (23/65) of genes related to nonresponse and activated 20% (16/79) of genes related to reaction (Fig. 5l). Seven of the 10 genes maximum strongly related to medical result have been regulated in a positive route. Conversely, most effective 4.9% (7/144) of genes have been regulated in a route opposing certain medical reaction, offering additional proof that BATF3 OE drives a transcriptional program related to certain medical results.CRISPRko monitors expose cofactors of BATF3BATF3 is a compact AP-1 TF with just a fundamental DNA binding area and a leucine zipper motif. For the reason that BATF3 lacks further protein domain names similar to transactivation domain names for gene activation, we speculated that BATF3 interacts with different TFs to have an effect on gene expression and chromatin accessibility (Supplementary Word 9)41. Moreover, we reasoned that different TFs may compete with or inhibit BATF3 and that disposing of those elements would additional enlarge the results of BATF3 OE. To spot those elements, we carried out parallel CRISPRko monitors without or with BATF3 OE the usage of a gRNA library concentrating on all 1,612 human TF genes45 (TFome) (Fig. 6a). We decided on IL7R expression because the readout for those monitors as a result of BATF3 OE profoundly will increase IL7R expression (Fig. 3a,b), thus offering a proxy for BATF3 task. IL7R could also be expressed in 20–50% of CD8+ T cells at baseline, making it possible to recuperate gene hits in each instructions, in contrast to ubiquitously silenced and extremely expressed genes.Fig. 6: CRISPRko monitors expose cofactors of BATF3 and different objectives for most cancers immunotherapy.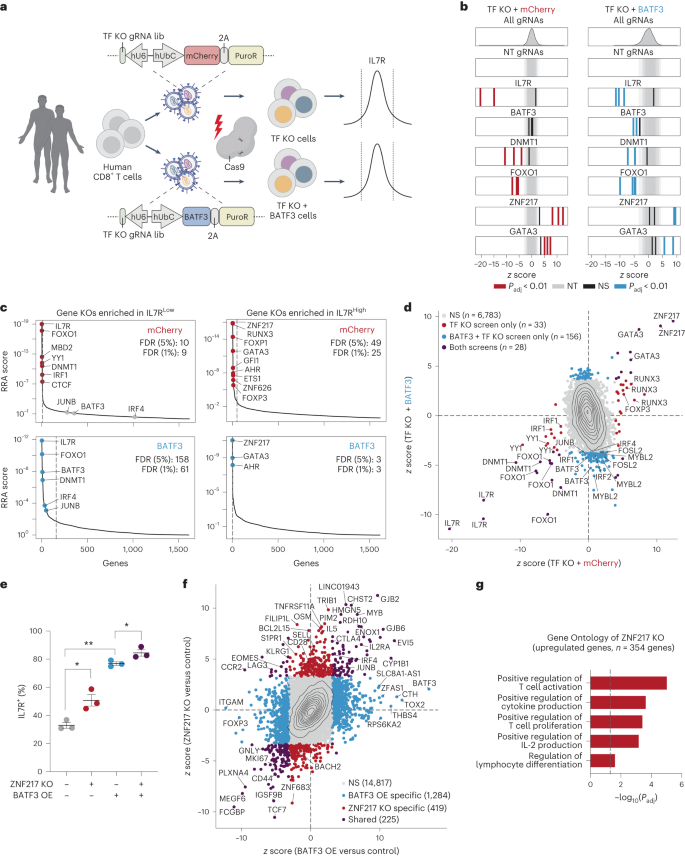 a, Schematic of CRISPRko monitors with TF KO gRNA library (lib). b, z ratings of gRNAs for decided on genes in mCherry (left) and BATF3 (proper) monitors. Enriched gRNAs (Padj < 0.01) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. c, Every gene goal within the mCherry (best) and BATF3 (backside) monitors ranked in keeping with the MAGeCK58 tough score aggregation (RRA) rating in each IL7RLOW (left) and IL7RHIGH (proper) populations. Dashed traces point out FDR of 0.05. d, Scatter plot of z ratings for each and every gRNA in CRISPRko monitors with BATF3 as opposed to with out BATF3. Enriched gRNAs (Padj < 0.01) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. e, Particular person and blended results of ZNF217 KO and BATF3 OE on IL7R expression (n = 3 donors, imply values ± s.e.m.). A one-way, paired ANOVA examine with Tukey’s put up hoc examine used to be used to match the share of IL7R+ cells between teams (Padj = 0.041 for keep watch over as opposed to ZNF217 KO, Padj = 0.008 for keep watch over as opposed to BATF3 OE, and Padj = 0.049 for BATF3 OE as opposed to BATF3 OE and ZNF217 KO). f, Scatter plot of transcriptomic results of ZNF217 KO as opposed to BATF3 OE relative to keep watch over T cells (n = 3 donors). DEGs (Padj < 0.05) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction and categorised at the foundation of whether or not the DEG used to be distinctive to a selected perturbation or shared throughout perturbations. g, Decided on enriched organic processes from ZNF217 KO. Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction.As anticipated, IL7R gRNAs have been essentially the most enriched gRNAs within the IL7R low inhabitants throughout each monitors (Fig. 6b). Significantly, BATF3 gRNAs most effective emerged within the display screen with BATF3 OE as BATF3 is lowly expressed at baseline (Fig. 6b). BATF3 gRNAs indiscriminately goal endogenous and exogenous BATF3, indicating that knocking out exogenous BATF3 nullified its results. Additional supporting the robustness of those monitors, we recovered more than one gRNA hits for lots of genes and the baseline expression of goal gene hits used to be considerably upper than non-hit genes (Prolonged Information Fig. 10a,b).Through evaluating gRNA- and gene-level enrichment between the 2 monitors (Fig. 6c,d), lets classify whether or not genes regulated IL7R in a BATF3-independent or BATF3-dependent means. As an example, FOXO1 and DNMT1 have been a few of the most powerful hits within the IL7R low inhabitants for each monitors, indicating BATF3-independent results. To spot attainable cofactors of BATF3, we looked for genes encoding for AP-1 or IRF TFs that have been most effective enriched within the IL7R low inhabitants with BATF3 OE. Significantly, BATF3, JUNB, and IRF4 have been the highest genes assembly those standards, confirming that BATF3 interacts with JUNB and IRF4 to mediate transcriptional keep watch over in CD8+ T cells (Fig. 6c and Prolonged Information Fig. 10c,d)46. Those monitors additionally printed upstream regulators of IL7R and candidate gene objectives for additional making improvements to ACT (Fig. 6c). Essentially the most enriched genes within the IL7R prime inhabitants within the TF-knockout (KO) display screen with out BATF3 OE have been ZNF217, RUNX3, FOXP1, GATA3, GFI1, AHR, ETS1, ZNF626 and FOXP3. Fewer genes have been enriched in IL7R prime inhabitants within the BATF3 OE display screen, partially as a result of baseline IL7R expression used to be upper. Moreover, we speculated that some TFs whose results have been misplaced with BATF3 OE could be downstream objectives of BATF3. Certainly, the RNA-seq effects display that a number of TFs together with FOXP1, ETS1 and FOXP3 have been all downregulated by means of BATF3 OE (Supplementary Desk 4).KO of 3 genes (ZNF217, GATA3 and AHR) larger IL7R expression for my part or together with BATF3 OE. ZNF217 used to be the highest hit in each monitors and has now not prior to now been characterised within the context of T mobile biology. GATA3 has been proven to advertise CD8+ T mobile disorder and focused deletion of GATA3 improves tumor control47. Additionally, each GATA3 and AHR can turn on FOXP3 expression in regulatory T cells, offering additional proof of a hyperlink between T mobile disorder and T mobile regulatory activity48,49,50.Subsequent, we measured the results of knocking out IL7R, BATF3, JUNB, IRF4, ZNF217 and GATA3 with and with out BATF3 OE (Prolonged Information Fig. 10e). BATF3 OE by myself larger IL7R expression by means of >40% in comparison to keep watch over CD8+ T cells (~33% to 77% IL7R+) (Prolonged Information Fig. 10e). Ablating BATF3 in part restored baseline IL7R ranges, possibly because of incomplete nuclease task throughout ectopic lentiviral copies of BATF3. IL7R induction by means of BATF3 used to be profoundly negated with both JUNB or IRF4 KOs (Prolonged Information Fig. 10e,f). Conversely, GATA3 and ZNF217 KOs larger the share of IL7R+ T cells (Prolonged Information Fig. 10e). In spite of everything, BATF3 OE and ZNF217 KO in combination resulted in an extra build up in T cells expressing IL7R (Fig. 6e and Prolonged Information Fig. 10g).We subsequent evaluated the transcriptional results of ZNF217 or GATA3 KO relative to keep watch over T cells and BATF3 OE by myself (Fig. 6f, Supplementary Fig. 11 and Supplementary Desk 7). ZNF217 KO resulted in 644 DEGs relative to keep watch over T cells with many encoding for TFs and floor makers implicated in T mobile biology and serve as (Fig. 6f). Additional supporting a T cell-specific function for ZNF217, Gene Ontology research printed that ZNF217 KO promoted certain law of T mobile activation, proliferation, IL-2 manufacturing, and differentiation (Fig. 6g and Supplementary Desk 7). Roughly 33% (225/644) of all DEGs with ZNF217 KO have been shared with BATF3 OE with the overwhelming majority (206/225) regulated in the similar route. However, the vast majority of DEGs for each and every particular person perturbation have been distinctive, suggesting that ZNF217 KO and BATF3 OE can power overlapping but in addition distinct transcriptional adjustments.
a, Schematic of CRISPRko monitors with TF KO gRNA library (lib). b, z ratings of gRNAs for decided on genes in mCherry (left) and BATF3 (proper) monitors. Enriched gRNAs (Padj < 0.01) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. c, Every gene goal within the mCherry (best) and BATF3 (backside) monitors ranked in keeping with the MAGeCK58 tough score aggregation (RRA) rating in each IL7RLOW (left) and IL7RHIGH (proper) populations. Dashed traces point out FDR of 0.05. d, Scatter plot of z ratings for each and every gRNA in CRISPRko monitors with BATF3 as opposed to with out BATF3. Enriched gRNAs (Padj < 0.01) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction. e, Particular person and blended results of ZNF217 KO and BATF3 OE on IL7R expression (n = 3 donors, imply values ± s.e.m.). A one-way, paired ANOVA examine with Tukey’s put up hoc examine used to be used to match the share of IL7R+ cells between teams (Padj = 0.041 for keep watch over as opposed to ZNF217 KO, Padj = 0.008 for keep watch over as opposed to BATF3 OE, and Padj = 0.049 for BATF3 OE as opposed to BATF3 OE and ZNF217 KO). f, Scatter plot of transcriptomic results of ZNF217 KO as opposed to BATF3 OE relative to keep watch over T cells (n = 3 donors). DEGs (Padj < 0.05) have been outlined the usage of a paired two-tailed DESeq2 examine with Benjamini–Hochberg correction and categorised at the foundation of whether or not the DEG used to be distinctive to a selected perturbation or shared throughout perturbations. g, Decided on enriched organic processes from ZNF217 KO. Statistical importance used to be outlined the usage of a two-tailed Fisher’s precise examine adopted by means of Benjamini–Hochberg correction.As anticipated, IL7R gRNAs have been essentially the most enriched gRNAs within the IL7R low inhabitants throughout each monitors (Fig. 6b). Significantly, BATF3 gRNAs most effective emerged within the display screen with BATF3 OE as BATF3 is lowly expressed at baseline (Fig. 6b). BATF3 gRNAs indiscriminately goal endogenous and exogenous BATF3, indicating that knocking out exogenous BATF3 nullified its results. Additional supporting the robustness of those monitors, we recovered more than one gRNA hits for lots of genes and the baseline expression of goal gene hits used to be considerably upper than non-hit genes (Prolonged Information Fig. 10a,b).Through evaluating gRNA- and gene-level enrichment between the 2 monitors (Fig. 6c,d), lets classify whether or not genes regulated IL7R in a BATF3-independent or BATF3-dependent means. As an example, FOXO1 and DNMT1 have been a few of the most powerful hits within the IL7R low inhabitants for each monitors, indicating BATF3-independent results. To spot attainable cofactors of BATF3, we looked for genes encoding for AP-1 or IRF TFs that have been most effective enriched within the IL7R low inhabitants with BATF3 OE. Significantly, BATF3, JUNB, and IRF4 have been the highest genes assembly those standards, confirming that BATF3 interacts with JUNB and IRF4 to mediate transcriptional keep watch over in CD8+ T cells (Fig. 6c and Prolonged Information Fig. 10c,d)46. Those monitors additionally printed upstream regulators of IL7R and candidate gene objectives for additional making improvements to ACT (Fig. 6c). Essentially the most enriched genes within the IL7R prime inhabitants within the TF-knockout (KO) display screen with out BATF3 OE have been ZNF217, RUNX3, FOXP1, GATA3, GFI1, AHR, ETS1, ZNF626 and FOXP3. Fewer genes have been enriched in IL7R prime inhabitants within the BATF3 OE display screen, partially as a result of baseline IL7R expression used to be upper. Moreover, we speculated that some TFs whose results have been misplaced with BATF3 OE could be downstream objectives of BATF3. Certainly, the RNA-seq effects display that a number of TFs together with FOXP1, ETS1 and FOXP3 have been all downregulated by means of BATF3 OE (Supplementary Desk 4).KO of 3 genes (ZNF217, GATA3 and AHR) larger IL7R expression for my part or together with BATF3 OE. ZNF217 used to be the highest hit in each monitors and has now not prior to now been characterised within the context of T mobile biology. GATA3 has been proven to advertise CD8+ T mobile disorder and focused deletion of GATA3 improves tumor control47. Additionally, each GATA3 and AHR can turn on FOXP3 expression in regulatory T cells, offering additional proof of a hyperlink between T mobile disorder and T mobile regulatory activity48,49,50.Subsequent, we measured the results of knocking out IL7R, BATF3, JUNB, IRF4, ZNF217 and GATA3 with and with out BATF3 OE (Prolonged Information Fig. 10e). BATF3 OE by myself larger IL7R expression by means of >40% in comparison to keep watch over CD8+ T cells (~33% to 77% IL7R+) (Prolonged Information Fig. 10e). Ablating BATF3 in part restored baseline IL7R ranges, possibly because of incomplete nuclease task throughout ectopic lentiviral copies of BATF3. IL7R induction by means of BATF3 used to be profoundly negated with both JUNB or IRF4 KOs (Prolonged Information Fig. 10e,f). Conversely, GATA3 and ZNF217 KOs larger the share of IL7R+ T cells (Prolonged Information Fig. 10e). In spite of everything, BATF3 OE and ZNF217 KO in combination resulted in an extra build up in T cells expressing IL7R (Fig. 6e and Prolonged Information Fig. 10g).We subsequent evaluated the transcriptional results of ZNF217 or GATA3 KO relative to keep watch over T cells and BATF3 OE by myself (Fig. 6f, Supplementary Fig. 11 and Supplementary Desk 7). ZNF217 KO resulted in 644 DEGs relative to keep watch over T cells with many encoding for TFs and floor makers implicated in T mobile biology and serve as (Fig. 6f). Additional supporting a T cell-specific function for ZNF217, Gene Ontology research printed that ZNF217 KO promoted certain law of T mobile activation, proliferation, IL-2 manufacturing, and differentiation (Fig. 6g and Supplementary Desk 7). Roughly 33% (225/644) of all DEGs with ZNF217 KO have been shared with BATF3 OE with the overwhelming majority (206/225) regulated in the similar route. However, the vast majority of DEGs for each and every particular person perturbation have been distinctive, suggesting that ZNF217 KO and BATF3 OE can power overlapping but in addition distinct transcriptional adjustments.
Transcriptional and epigenetic regulators of human CD8+ T mobile operate recognized via orthogonal CRISPR monitors – Nature Genetics